+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0333 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical RNA-bound Hantaan virus nucleocapsid | |||||||||
 Map data Map data | Helical RNA-bound Hantaan virus nucleocapsid structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleoprotein / Nucleocapsid / RNA-binding / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host apoptosis / helical viral capsid / viral nucleocapsid / endonuclease activity / host cell Golgi apparatus / Hydrolases; Acting on ester bonds / host cell perinuclear region of cytoplasm / ribonucleoprotein complex / hydrolase activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Hantaan orthohantavirus / Hantaan orthohantavirus /  | |||||||||
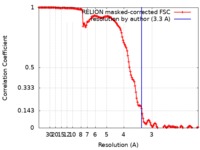
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Arragain B / Reguera J | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms. Authors: Benoît Arragain / Juan Reguera / Ambroise Desfosses / Irina Gutsche / Guy Schoehn / Hélène Malet /  Abstract: Negative-strand RNA viruses condense their genome into helical nucleocapsids that constitute essential templates for viral replication and transcription. The intrinsic flexibility of nucleocapsids ...Negative-strand RNA viruses condense their genome into helical nucleocapsids that constitute essential templates for viral replication and transcription. The intrinsic flexibility of nucleocapsids usually prevents their full-length structural characterisation at high resolution. Here, we describe purification of full-length recombinant metastable helical nucleocapsid of Hantaan virus ( family, order) and determine its structure at 3.3 Å resolution by cryo-electron microscopy. The structure reveals the mechanisms of helical multimerisation via sub-domain exchanges between protomers and highlights nucleotide positions in a continuous positively charged groove compatible with viral genome binding. It uncovers key sites for future structure-based design of antivirals that are currently lacking to counteract life-threatening hantavirus infections. The structure also suggests a model of nucleoprotein-polymerase interaction that would enable replication and transcription solely upon local disruption of the nucleocapsid. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0333.map.gz emd_0333.map.gz | 14 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0333-v30.xml emd-0333-v30.xml emd-0333.xml emd-0333.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0333_fsc.xml emd_0333_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0333.png emd_0333.png | 149.2 KB | ||
| Filedesc metadata |  emd-0333.cif.gz emd-0333.cif.gz | 7.3 KB | ||
| Others |  emd_0333_half_map_1.map.gz emd_0333_half_map_1.map.gz emd_0333_half_map_2.map.gz emd_0333_half_map_2.map.gz | 228.7 MB 228.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0333 http://ftp.pdbj.org/pub/emdb/structures/EMD-0333 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0333 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0333 | HTTPS FTP |
-Related structure data
| Related structure data |  6i2nMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0333.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0333.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical RNA-bound Hantaan virus nucleocapsid structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 2
| File | emd_0333_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_0333_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RNA-bound Hantaan virus nucleocapsid
| Entire | Name: RNA-bound Hantaan virus nucleocapsid |
|---|---|
| Components |
|
-Supramolecule #1: RNA-bound Hantaan virus nucleocapsid
| Supramolecule | Name: RNA-bound Hantaan virus nucleocapsid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 48.2 KDa |
-Supramolecule #2: Hantaan virus nucleoprotein
| Supramolecule | Name: Hantaan virus nucleoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Hantaan orthohantavirus Hantaan orthohantavirus |
-Supramolecule #3: RNA
| Supramolecule | Name: RNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RNA (5'-R(P*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*U)-3') / type: rna / ID: 1 Details: A poly-U has been built but the RNA corresponds to heterogeneous insect cell RNA that bind to nucleoproteins during expression. Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 873.54 Da |
| Sequence | String: UUU |
-Macromolecule #2: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hantaan orthohantavirus Hantaan orthohantavirus |
| Molecular weight | Theoretical: 48.262266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GMATMEELQR EINAHEGQLV IARQKVRDAE KQYEKDPDEL NKRTLTDREG VAVSIQAKID ELKRQLADRI ATGKNLGKEQ DPTGVEPGD HLKERSMLSY GNVLDLNHLD IDEPTGQTAD WLSIIVYLTS FVVPILLKAL YMLTTRGRQT TKDNKGTRIR F KDDSSFED ...String: GMATMEELQR EINAHEGQLV IARQKVRDAE KQYEKDPDEL NKRTLTDREG VAVSIQAKID ELKRQLADRI ATGKNLGKEQ DPTGVEPGD HLKERSMLSY GNVLDLNHLD IDEPTGQTAD WLSIIVYLTS FVVPILLKAL YMLTTRGRQT TKDNKGTRIR F KDDSSFED VNGIRKPKHL YVSLPNAQSS MKAEEITPGR YRTAVCGLYP AQIKARQMIS PVMSVIGFLA LAKDWSDRIE QW LIEPCKL LPDTAAVSLL GGPATNRDYL RQRQVALGNM ETKESKAIRQ HAEAAGCSMI EDIESPSSIW VFAGAPDRCP PTC LFIAGI AELGAFFSIL QDMRNTIMAS KTVGTSEEKL RKKSSFYQSY LRRTQSMGIQ LGQRIIVLFM VAWGKEAVDN FHLG DDMDP ELRTLAQSLI DVKVKEISNQ EPLKL UniProtKB: Nucleoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: 30 mA | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV Details: 3 ul of sample were applied on the resulting glow-discharged grids and excess solution was blotted during 2.5 sec force 7. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3840 pixel / Digitization - Dimensions - Height: 3712 pixel / Digitization - Frames/image: 3-18 / Number grids imaged: 1 / Number real images: 4328 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.5 µm / Calibrated defocus min: 0.8 µm / Calibrated magnification: 46860 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Residue range: 113-429 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Fiber model was iteratively rebuilt and all-atom refined using stereochemical and NCS restraints |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 103 |
| Output model |  PDB-6i2n: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)