+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0327 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vibrio vulnificus EpsD | ||||||||||||
 Map data Map data | EpsD 3D reconstruction | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | PROTEIN TRANSPORT | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein secretion by the type II secretion system / type II protein secretion system complex / membrane => GO:0016020 / cell outer membrane Similarity search - Function | ||||||||||||
| Biological species |  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) | ||||||||||||
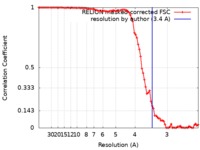
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Contreras-Martel C / Farias Estrozi L | ||||||||||||
| Funding support |  France, France,  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2019 Journal: PLoS Pathog / Year: 2019Title: Structure and assembly of pilotin-dependent and -independent secretins of the type II secretion system. Authors: S Peter Howard / Leandro F Estrozi / Quentin Bertrand / Carlos Contreras-Martel / Timothy Strozen / Viviana Job / Alexandre Martins / Daphna Fenel / Guy Schoehn / Andréa Dessen /    Abstract: The type II secretion system (T2SS) is a cell envelope-spanning macromolecular complex that is prevalent in Gram-negative bacterial species. It serves as the predominant virulence mechanism of many ...The type II secretion system (T2SS) is a cell envelope-spanning macromolecular complex that is prevalent in Gram-negative bacterial species. It serves as the predominant virulence mechanism of many bacteria including those of the emerging human pathogens Vibrio vulnificus and Aeromonas hydrophila. The system is composed of a core set of highly conserved proteins that assemble an inner membrane platform, a periplasmic pseudopilus and an outer membrane complex termed the secretin. Localization and assembly of secretins in the outer membrane requires recognition of secretin monomers by two different partner systems: an inner membrane accessory complex or a highly sequence-diverse outer membrane lipoprotein, termed the pilotin. In this study, we addressed the question of differential secretin assembly mechanisms by using cryo-electron microscopy to determine the structures of the secretins from A. hydrophila (pilotin-independent ExeD) and V. vulnificus (pilotin-dependent EpsD). These structures, at approximately 3.5 Å resolution, reveal pentadecameric stoichiometries and C-terminal regions that carry a signature motif in the case of a pilotin-dependent assembly mechanism. We solved the crystal structure of the V. vulnificus EpsS pilotin and confirmed the importance of the signature motif for pilotin-dependent secretin assembly by performing modelling with the C-terminus of EpsD. We also show that secretin assembly is essential for membrane integrity and toxin secretion in V. vulnificus and establish that EpsD requires the coordinated activity of both the accessory complex EpsAB and the pilotin EpsS for full assembly and T2SS function. In contrast, mutation of the region of the S-domain that is normally the site of pilotin interactions has little effect on assembly or function of the ExeD secretin. Since secretins are essential outer membrane channels present in a variety of secretion systems, these results provide a structural and functional basis for understanding the key assembly steps for different members of this vast pore-forming family of proteins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0327.map.gz emd_0327.map.gz | 10.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0327-v30.xml emd-0327-v30.xml emd-0327.xml emd-0327.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0327_fsc.xml emd_0327_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0327.png emd_0327.png | 221.1 KB | ||
| Masks |  emd_0327_msk_1.map emd_0327_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0327.cif.gz emd-0327.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0327 http://ftp.pdbj.org/pub/emdb/structures/EMD-0327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0327 | HTTPS FTP |
-Related structure data
| Related structure data |  6i1yMC  0326C  6i1xC  6i2vC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0327.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0327.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EpsD 3D reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0327_msk_1.map emd_0327_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : EpsD
| Entire | Name: EpsD |
|---|---|
| Components |
|
-Supramolecule #1: EpsD
| Supramolecule | Name: EpsD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) |
| Molecular weight | Theoretical: 1 MDa |
-Macromolecule #1: General secretion pathway protein GspD
| Macromolecule | Name: General secretion pathway protein GspD / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio vulnificus (bacteria) Vibrio vulnificus (bacteria) |
| Molecular weight | Theoretical: 60.124922 KDa |
| Recombinant expression | Organism: Cell-free gateway cloning vector N-term 8xHis pCellFree_G01 (others) |
| Sequence | String: DNVITRVVAV RNVSVRELSP LLRQLIDNAG AGNVVHYDPA NIILITGRAA VVNRLAEIIK RVDQAGNREI EVVELGNASA AEMVRIVDA LNRTTDAKNT PEFLQPKLVA DERTNSILIS GDPKVRDRLK RLIRQLDVEM ASKGNNRVVY LKYAKAEDLV D VLKGVSDN ...String: DNVITRVVAV RNVSVRELSP LLRQLIDNAG AGNVVHYDPA NIILITGRAA VVNRLAEIIK RVDQAGNREI EVVELGNASA AEMVRIVDA LNRTTDAKNT PEFLQPKLVA DERTNSILIS GDPKVRDRLK RLIRQLDVEM ASKGNNRVVY LKYAKAEDLV D VLKGVSDN LQAEKNSGQK GASSQRNDVV IAAHQGTNSL VLTAPPDIML ALQEVITQLD IRRAQVLIEA LIVEMAEGDG VN LGVQWGN LETGAVIQYS NTGTPIGKVM VGLEEAKDKT VTDSRWNSDT DKYEPYSRTE AGDYSTLAAA LAGVNGAAMS LVM GDWTAL ISAVSSDSNS NILSSPSITV MDNGEASFIV GEEVPVITGS TAGSNNDNPF QTVDRKEVGI KLKVVPQINE GDSV QLNIE QEVSNVLGAN GAVDVRFAKR QLNTSVIVQD GQMLVLGGLI DERALESESK VPLLGDIPIL GHLFKSTNTQ VEKKN LMVF IKPTIIRDGM TADGITQRKY NYIRAEQLYK AEQGLKLMDD GHIPVLPKFG EDKRHPAEIQ AFIDQMEQQ UniProtKB: General secretion pathway protein GspD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 2021 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 41322 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Coot and buccaneer for model building and refmac5 for refinement (CCPEM-1.1.0 suite). |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 146 / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-6i1y: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)