[English] 日本語
 Yorodumi
Yorodumi- EMDB-0268: Complex IV in the III2-IV2 mitochondrial respiratory supercomplex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0268 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex IV in the III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae (CIVb) | |||||||||
 Map data Map data | The second complex IV of the III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
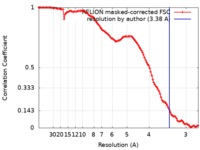
| Method | single particle reconstruction / cryo EM / Resolution: 3.38 Å | |||||||||
 Authors Authors | Hartley AM / Lukoyanova N / Pinotsis N / Marechal A | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc. Authors: Andrew M Hartley / Natalya Lukoyanova / Yunyi Zhang / Alfredo Cabrera-Orefice / Susanne Arnold / Brigitte Meunier / Nikos Pinotsis / Amandine Maréchal /    Abstract: Cytochrome c oxidase (complex IV, CIV) is known in mammals to exist independently or in association with other respiratory proteins to form supercomplexes (SCs). In Saccharomyces cerevisiae, CIV is ...Cytochrome c oxidase (complex IV, CIV) is known in mammals to exist independently or in association with other respiratory proteins to form supercomplexes (SCs). In Saccharomyces cerevisiae, CIV is found solely in an SC with cytochrome bc (complex III, CIII). Here, we present the cryogenic electron microscopy (cryo-EM) structure of S. cerevisiae CIV in a IIIIV SC at 3.3 Å resolution. While overall similarity to mammalian homologs is high, we found notable differences in the supernumerary subunits Cox26 and Cox13; the latter exhibits a unique arrangement that precludes CIV dimerization as seen in bovine. A conformational shift in the matrix domain of Cox5A-involved in allosteric inhibition by ATP-may arise from its association with CIII. The CIII-CIV arrangement highlights a conserved interaction interface of CIII, albeit one occupied by complex I in mammalian respirasomes. We discuss our findings in the context of the potential impact of SC formation on CIV regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0268.map.gz emd_0268.map.gz | 99.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0268-v30.xml emd-0268-v30.xml emd-0268.xml emd-0268.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0268_fsc.xml emd_0268_fsc.xml | 11 KB | Display |  FSC data file FSC data file |
| Images |  emd_0268.png emd_0268.png | 247.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0268 http://ftp.pdbj.org/pub/emdb/structures/EMD-0268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0268 | HTTPS FTP |
-Validation report
| Summary document |  emd_0268_validation.pdf.gz emd_0268_validation.pdf.gz | 306 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0268_full_validation.pdf.gz emd_0268_full_validation.pdf.gz | 305.1 KB | Display | |
| Data in XML |  emd_0268_validation.xml.gz emd_0268_validation.xml.gz | 11.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0268 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0268 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0268 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0268 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0268.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0268.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second complex IV of the III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3861 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
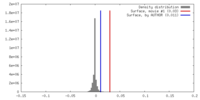
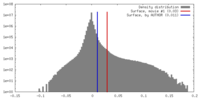
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : 12-subunit cytochrome c oxidase with isoform Cox5A
+Supramolecule #1: 12-subunit cytochrome c oxidase with isoform Cox5A
+Macromolecule #1: Cox1
+Macromolecule #2: Cox2
+Macromolecule #3: Cox3
+Macromolecule #4: Cox4
+Macromolecule #5: Cox5A
+Macromolecule #6: Cox6
+Macromolecule #7: Cox7
+Macromolecule #8: Cox8
+Macromolecule #9: Cox9
+Macromolecule #10: Cox12
+Macromolecule #11: Cox13
+Macromolecule #13: Cox26
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 92 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL of sample applied to negatively glow discharged grid, blot force -10; blotting time 8.5 sec. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Sampling interval: 5.0 µm / Average exposure time: 8.0 sec. / Average electron dose: 1.645 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)