[English] 日本語
 Yorodumi
Yorodumi- EMDB-0139: Structure of a hibernating 100S ribosome reveals an inactive conf... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0139 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 100S / cryo-EM / E-site tRNA / hibernation / HPF / ribosome / RMF / S1 | |||||||||
| Function / homology |  Function and homology information Function and homology informationdormancy process / negative regulation of translation in response to stress / negative regulation of translational elongation / positive regulation of cytoplasmic translation / cellular response to stress / negative regulation of cytoplasmic translational initiation / transcription antitermination factor activity, RNA binding / ornithine decarboxylase inhibitor activity / ribosomal small subunit binding / misfolded RNA binding ...dormancy process / negative regulation of translation in response to stress / negative regulation of translational elongation / positive regulation of cytoplasmic translation / cellular response to stress / negative regulation of cytoplasmic translational initiation / transcription antitermination factor activity, RNA binding / ornithine decarboxylase inhibitor activity / ribosomal small subunit binding / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity / negative regulation of cytoplasmic translation / four-way junction DNA binding / DnaA-L2 complex / translation repressor activity / negative regulation of translational initiation / regulation of mRNA stability / negative regulation of DNA-templated DNA replication initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / response to reactive oxygen species / regulation of DNA-templated transcription elongation / ribosome assembly / transcription elongation factor complex / transcription antitermination / DNA endonuclease activity / regulation of cell growth / translational initiation / DNA-templated transcription termination / response to radiation / maintenance of translational fidelity / mRNA 5'-UTR binding / regulation of translation / large ribosomal subunit / ribosome biogenesis / transferase activity / ribosome binding / ribosomal small subunit assembly / ribosomal small subunit biogenesis / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / single-stranded RNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / hydrolase activity / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA binding / RNA binding / zinc ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.9 Å | |||||||||
 Authors Authors | Beckert B / Turk M | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2018 Journal: Nat Microbiol / Year: 2018Title: Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Authors: Bertrand Beckert / Martin Turk / Andreas Czech / Otto Berninghausen / Roland Beckmann / Zoya Ignatova / Jürgen M Plitzko / Daniel N Wilson /  Abstract: To survive under conditions of stress, such as nutrient deprivation, bacterial 70S ribosomes dimerize to form hibernating 100S particles. In γ-proteobacteria, such as Escherichia coli, 100S ...To survive under conditions of stress, such as nutrient deprivation, bacterial 70S ribosomes dimerize to form hibernating 100S particles. In γ-proteobacteria, such as Escherichia coli, 100S formation requires the ribosome modulation factor (RMF) and the hibernation promoting factor (HPF). Here we present single-particle cryo-electron microscopy structures of hibernating 70S and 100S particles isolated from stationary-phase E. coli cells at 3.0 Å and 7.9 Å resolution, respectively. The structures reveal the binding sites for HPF and RMF as well as the unexpected presence of deacylated E-site transfer RNA and ribosomal protein bS1. HPF interacts with the anticodon-stem-loop of the E-tRNA and occludes the binding site for the messenger RNA as well as A- and P-site tRNAs. RMF facilitates stabilization of a compact conformation of bS1, which together sequester the anti-Shine-Dalgarno sequence of the 16S ribosomal RNA (rRNA), thereby inhibiting translation initiation. At the dimerization interface, the C-terminus of uS2 probes the mRNA entrance channel of the symmetry-related particle, thus suggesting that dimerization inactivates ribosomes by blocking the binding of mRNA within the channel. The back-to-back E. coli 100S arrangement is distinct from 100S particles observed previously in Gram-positive bacteria, and reveals a unique role for bS1 in translation regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0139.map.gz emd_0139.map.gz | 3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0139-v30.xml emd-0139-v30.xml emd-0139.xml emd-0139.xml | 72 KB 72 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0139_fsc.xml emd_0139_fsc.xml | 8.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0139.png emd_0139.png | 174 KB | ||
| Filedesc metadata |  emd-0139.cif.gz emd-0139.cif.gz | 13.6 KB | ||
| Others |  emd_0139_additional_1.map.gz emd_0139_additional_1.map.gz emd_0139_additional_2.map.gz emd_0139_additional_2.map.gz | 286.9 MB 286.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0139 http://ftp.pdbj.org/pub/emdb/structures/EMD-0139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0139 | HTTPS FTP |
-Related structure data
| Related structure data |  6h58MC  0137C  6h4nC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0139.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0139.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
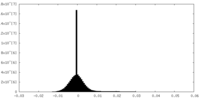
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_0139_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
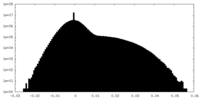
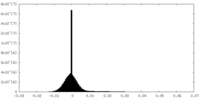
| Density Histograms |
-Additional map: #2
| File | emd_0139_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
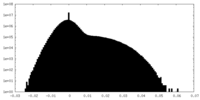
| Density Histograms |
- Sample components
Sample components
+Entire : Structure of a hibernating 100S ribosome reveals an inactive conf...
+Supramolecule #1: Structure of a hibernating 100S ribosome reveals an inactive conf...
+Macromolecule #1: 50S ribosomal protein L32
+Macromolecule #2: 50S ribosomal protein L33
+Macromolecule #3: 50S ribosomal protein L34
+Macromolecule #4: 50S ribosomal protein L35
+Macromolecule #5: 50S ribosomal protein L36
+Macromolecule #6: 50S ribosomal protein L31
+Macromolecule #9: 50S ribosomal protein L2
+Macromolecule #10: 50S ribosomal protein L3
+Macromolecule #11: 50S ribosomal protein L4
+Macromolecule #12: 50S ribosomal protein L5
+Macromolecule #13: 50S ribosomal protein L6
+Macromolecule #14: 50S ribosomal protein L9
+Macromolecule #15: 50S ribosomal protein L13
+Macromolecule #16: 50S ribosomal protein L14
+Macromolecule #17: 50S ribosomal protein L15
+Macromolecule #18: 50S ribosomal protein L16
+Macromolecule #19: 50S ribosomal protein L17
+Macromolecule #20: 50S ribosomal protein L18
+Macromolecule #21: 50S ribosomal protein L19
+Macromolecule #22: 50S ribosomal protein L20
+Macromolecule #23: 50S ribosomal protein L21
+Macromolecule #24: 50S ribosomal protein L22
+Macromolecule #25: 50S ribosomal protein L23
+Macromolecule #26: 50S ribosomal protein L24
+Macromolecule #27: 50S ribosomal protein L25
+Macromolecule #28: 50S ribosomal protein L27
+Macromolecule #29: 50S ribosomal protein L28
+Macromolecule #30: 50S ribosomal protein L29
+Macromolecule #31: 50S ribosomal protein L30
+Macromolecule #33: 30S ribosomal protein S2
+Macromolecule #34: 30S ribosomal protein S3
+Macromolecule #35: 30S ribosomal protein S4
+Macromolecule #36: 30S ribosomal protein S5
+Macromolecule #37: 30S ribosomal protein S6
+Macromolecule #38: 30S ribosomal protein S7
+Macromolecule #39: 30S ribosomal protein S8
+Macromolecule #40: 30S ribosomal protein S9
+Macromolecule #41: 30S ribosomal protein S10
+Macromolecule #42: 30S ribosomal protein S11
+Macromolecule #43: 30S ribosomal protein S12
+Macromolecule #44: 30S ribosomal protein S13
+Macromolecule #45: 30S ribosomal protein S14
+Macromolecule #46: 30S ribosomal protein S15
+Macromolecule #47: 30S ribosomal protein S16
+Macromolecule #48: 30S ribosomal protein S17
+Macromolecule #49: 30S ribosomal protein S18
+Macromolecule #50: 30S ribosomal protein S19
+Macromolecule #51: 30S ribosomal protein S20
+Macromolecule #52: 30S ribosomal protein S21
+Macromolecule #53: Ribosome modulation factor
+Macromolecule #54: Ribosome hibernation promoting factor
+Macromolecule #55: 30S ribosomal protein S1
+Macromolecule #7: 23S ribosomal RNA
+Macromolecule #8: 5S ribosomal RNA
+Macromolecule #32: 16S ribosomal RNA
+Macromolecule #56: tRNA Mixture
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.15 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)