+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
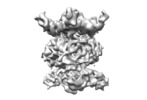
| Title | MlaBDEF complex from A. baumannii | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationphospholipid transporter activity / membrane => GO:0016020 / ATP-binding cassette (ABC) transporter complex / ATP hydrolysis activity / ATP binding Similarity search - Function | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.7 Å | |||||||||
 Authors Authors | Bergeron JRC / Kollman JM | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: The Mla system and glycerophospholipid transport to the outer membrane. Authors: Cassandra Kamischke / Junping Fan / Julien Bergeron / Hemantha D Kulasekara / Zachary D Dalebroux / Anika Burrell / Justin M Kollman / Samuel I Miller /   Abstract: The outer membrane (OM) of Gram-negative bacteria serves as a selective permeability barrier that allows entry of essential nutrients while excluding toxic compounds, including antibiotics. The OM is ...The outer membrane (OM) of Gram-negative bacteria serves as a selective permeability barrier that allows entry of essential nutrients while excluding toxic compounds, including antibiotics. The OM is asymmetric and contains an outer leaflet of lipopolysaccharides (LPS) or lipooligosaccharides (LOS) and an inner leaflet of glycerophospholipids (GPL). We screened transposon mutants and identified a number of mutants with OM defects, including an ABC transporter system homologous to the Mla system in We further show that this opportunistic, antibiotic-resistant pathogen uses this multicomponent protein complex and ATP hydrolysis at the inner membrane to promote GPL export to the OM. The broad conservation of the Mla system in Gram-negative bacteria suggests the system may play a conserved role in OM biogenesis. The importance of the Mla system to OM integrity and antibiotic sensitivity suggests that its components may serve as new antimicrobial therapeutic targets. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0007.map.gz emd_0007.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0007-v30.xml emd-0007-v30.xml emd-0007.xml emd-0007.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0007.png emd_0007.png | 64.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0007 http://ftp.pdbj.org/pub/emdb/structures/EMD-0007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0007 | HTTPS FTP |
-Related structure data
| Related structure data |  6ic4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0007.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0007.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.26 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : MlaBDEF
| Entire | Name: MlaBDEF |
|---|---|
| Components |
|
-Supramolecule #1: MlaBDEF
| Supramolecule | Name: MlaBDEF / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #1: MlaB
| Macromolecule | Name: MlaB / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MVQYLNQELV VSGKIDFENA EQQYQAGLAI IKKQTSFPLI VDLKQLEHGN TLALAVLVQW LRQTPQKSGL HFKNVPEKML KIIQACHLQ EDLHLV |
-Macromolecule #2: MlaD
| Macromolecule | Name: MlaD / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MKSRTSELAV GIFVIIFGIA LFFLAMKVSG LVGTNLSDGY TMKAQFDNVN GLKPRAKVTM SGVTIGRVDS ITLDPVTRLA TVTFDLDGK LTSFNAEQLK EVQKNALDEL RYSSDYTQAT PAQQKTMEQQ LISNMNSITS IDEDAYIMVA TNGLLGEKYL K IVPGGGLN ...String: MKSRTSELAV GIFVIIFGIA LFFLAMKVSG LVGTNLSDGY TMKAQFDNVN GLKPRAKVTM SGVTIGRVDS ITLDPVTRLA TVTFDLDGK LTSFNAEQLK EVQKNALDEL RYSSDYTQAT PAQQKTMEQQ LISNMNSITS IDEDAYIMVA TNGLLGEKYL K IVPGGGLN YLKRGDTISN TQGTMDLEDL ISKFITGGGA GKVAAGSSSA EEKAPASTDS SAQPSFVE |
-Macromolecule #3: MlaE
| Macromolecule | Name: MlaE / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MNTIAWLGRL VIERIRGIGV AALMLLQIIF SLPSAGGFGR FVYQMHRVGV MSLLIITVSG LFIGLVLGLQ GYSILVNVGS ESMLGTMVS LTLLRELAPV VAALLFAGRA GSALTAEIGS MKQSEQLASM EMIGVDPLKQ IVSPRLWAGI VSLPMLTVIF A AIGIVGGK ...String: MNTIAWLGRL VIERIRGIGV AALMLLQIIF SLPSAGGFGR FVYQMHRVGV MSLLIITVSG LFIGLVLGLQ GYSILVNVGS ESMLGTMVS LTLLRELAPV VAALLFAGRA GSALTAEIGS MKQSEQLASM EMIGVDPLKQ IVSPRLWAGI VSLPMLTVIF A AIGIVGGK LVGVDFLGVD EGSFWSGMQN NVQFGHDVVN GIIKSIVFAL LCTWIAVFQG YACDPTPEGI ATAMTRTVVY SS LCVLGFD FVLTAVMFGG I |
-Macromolecule #4: MlaF
| Macromolecule | Name: MlaF / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Sequence | String: MIAIMNNKTP LSTQSLIEVK NLSFNRGERV IYDNISLNIR RGQITAIMGP SGTGKTTLLR LIGGQLVPDQ GEVLLDGKDI AQMSRQELF AARARMGMLF QSGALFTDMS VYENVAFPIR AHTKLSENLI AELVALKLES VGLRGTEQLM PTELSGGMNR R VALARAIA ...String: MIAIMNNKTP LSTQSLIEVK NLSFNRGERV IYDNISLNIR RGQITAIMGP SGTGKTTLLR LIGGQLVPDQ GEVLLDGKDI AQMSRQELF AARARMGMLF QSGALFTDMS VYENVAFPIR AHTKLSENLI AELVALKLES VGLRGTEQLM PTELSGGMNR R VALARAIA LDPDLIMYDE PFAGQDPIVK GVLTRLIRSL REALDLTTII VSHDVPETLS IADYIYVVAE GKIQGEGTPE EL QAYASPF VKQFLTGSAE GPVEYQFSHQ AYLDNEVRP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 8.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 14000 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















