+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDER5 |
|---|---|
 Sample Sample | AICAR/fumarate-bound neanderthal adenylosuccinate lyase (ADSL)
|
| Biological species |  Homo sapiens neanderthalensis (Neandertal) Homo sapiens neanderthalensis (Neandertal) |
 Citation Citation |  Journal: Sci Rep / Year: 2018 Journal: Sci Rep / Year: 2018Title: Molecular comparison of Neanderthal and Modern Human adenylosuccinate lyase. Authors: Bart Van Laer / Ulrike Kapp / Montserrat Soler-Lopez / Kaja Moczulska / Svante Pääbo / Gordon Leonard / Christoph Mueller-Dieckmann /    Abstract: The availability of genomic data from extinct homini such as Neanderthals has caused a revolution in palaeontology allowing the identification of modern human-specific protein substitutions. ...The availability of genomic data from extinct homini such as Neanderthals has caused a revolution in palaeontology allowing the identification of modern human-specific protein substitutions. Currently, little is known as to how these substitutions alter the proteins on a molecular level. Here, we investigate adenylosuccinate lyase, a conserved enzyme involved in purine metabolism for which several substitutions in the modern human protein (hADSL) have been described to affect intelligence and behaviour. During evolution, modern humans acquired a specific substitution (Ala429Val) in ADSL distinguishing it from the ancestral variant present in Neanderthals (nADSL). We show here that despite this conservative substitution being solvent exposed and located distant from the active site, there is a difference in thermal stability, but not enzymology or ligand binding between nADSL and hADSL. Substitutions near residue 429 which do not profoundly affect enzymology were previously reported to cause neurological symptoms in humans. This study also reveals that ADSL undergoes conformational changes during catalysis which, together with the crystal structure of a hitherto undetermined product bound conformation, explains the molecular origin of disease for several modern human ADSL mutants. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #2432 |  Type: atomic / Chi-square value: 11.538  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: AICAR/fumarate-bound neanderthal adenylosuccinate lyase (ADSL) Specimen concentration: 5 mg/ml |
|---|---|
| Buffer | Name: 10 mM HEPES, 100 mM NaCl, 1 mM DTT, 1 mM 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), 1mM Fumarate pH: 7.5 |
| Entity #1311 | Name: ADSL / Type: protein / Description: Adenylosuccinate Lyase / Formula weight: 55.142 / Num. of mol.: 4 / Source: Homo sapiens neanderthalensis Sequence: GSHMAAGGDH GSPDSYRSPL ASRYASPEMC FVFSDRYKFR TWRQLWLWLA EAEQTLGLPI TDEQIQEMKS NLENIDFKMA AEEEKRLRHD VMAHVHTFGH CCPKAAGIIH LGATSCYVGD NTDLIILRNA LDLLLPKLAR VISRLADFAK ERASLPTLGF THFQPAQLTT ...Sequence: GSHMAAGGDH GSPDSYRSPL ASRYASPEMC FVFSDRYKFR TWRQLWLWLA EAEQTLGLPI TDEQIQEMKS NLENIDFKMA AEEEKRLRHD VMAHVHTFGH CCPKAAGIIH LGATSCYVGD NTDLIILRNA LDLLLPKLAR VISRLADFAK ERASLPTLGF THFQPAQLTT VGKRCCLWIQ DLCMDLQNLK RVRDDLRFRG VKGTTGTQAS FLQLFEGDDH KVEQLDKMVT EKAGFKRAFI ITGQTYTRKV DIEVLSVLAS LGASVHKICT DIRLLANLKE MEEPFEKQQI GSSAMPYKRN PMRSERCCSL ARHLMTLVMD PLQTASVQWF ERTLDDSANR RICLAEAFLT ADTILNTLQN ISEGLVVYPK VIERRIRQEL PFMATENIIM AMVKAGGSRQ DCHEKIRVLS QQAASVVKQE GGDNDLIERI QADAYFSPIH SQLDHLLDPS SFTGRASQQV QRFLEEEVYP LLKPYESVMK VKAELCL |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.864 mm / Type of source: X-ray synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.864 mm | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | ||||||||||||||||||||||||
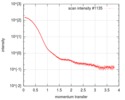
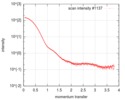
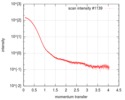
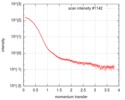
| Scan | Measurement date: Jul 24, 2016 / Storage temperature: 20 °C / Cell temperature: 20 °C / Exposure time: 1 sec. / Number of frames: 10 / Unit: 1/nm /
| ||||||||||||||||||||||||
| Result | Type of curve: single_conc /
|
 Movie
Movie Controller
Controller


 SASDER5
SASDER5