[English] 日本語
 Yorodumi
Yorodumi- SASDCY8: Human Rev7 monomer (R124A mutant) in complex with Rev3 peptide @ ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDCY8 |
|---|---|
 Sample Sample | Human Rev7 monomer (R124A mutant) in complex with Rev3 peptide @ 8mg/mL
|
| Function / homology |  Function and homology information Function and homology informationsomatic diversification of immunoglobulins involved in immune response / DNA damage response, signal transduction resulting in transcription / zeta DNA polymerase complex / positive regulation of isotype switching / negative regulation of transcription by competitive promoter binding / negative regulation of cell-cell adhesion mediated by cadherin / JUN kinase binding / negative regulation of epithelial to mesenchymal transition / negative regulation of ubiquitin protein ligase activity / positive regulation of double-strand break repair via nonhomologous end joining ...somatic diversification of immunoglobulins involved in immune response / DNA damage response, signal transduction resulting in transcription / zeta DNA polymerase complex / positive regulation of isotype switching / negative regulation of transcription by competitive promoter binding / negative regulation of cell-cell adhesion mediated by cadherin / JUN kinase binding / negative regulation of epithelial to mesenchymal transition / negative regulation of ubiquitin protein ligase activity / positive regulation of double-strand break repair via nonhomologous end joining / mitotic spindle assembly checkpoint signaling / telomere maintenance in response to DNA damage / positive regulation of peptidyl-serine phosphorylation / error-prone translesion synthesis / negative regulation of double-strand break repair via homologous recombination / actin filament organization / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / regulation of cell growth / negative regulation of canonical Wnt signaling pathway / double-strand break repair via homologous recombination / negative regulation of protein catabolic process / DNA-templated DNA replication / spindle / transcription corepressor activity / double-strand break repair / site of double-strand break / chromosome / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-directed DNA polymerase activity / cell division / nucleotide binding / chromatin / positive regulation of DNA-templated transcription / nucleolus / negative regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex. Authors: Alessandro A Rizzo / Faye-Marie Vassel / Nimrat Chatterjee / Sanjay D'Souza / Yunfeng Li / Bing Hao / Michael T Hemann / Graham C Walker / Dmitry M Korzhnev /  Abstract: The translesion synthesis (TLS) polymerases Polζ and Rev1 form a complex that enables replication of damaged DNA. The Rev7 subunit of Polζ, which is a multifaceted HORMA (Hop1, Rev7, Mad2) protein ...The translesion synthesis (TLS) polymerases Polζ and Rev1 form a complex that enables replication of damaged DNA. The Rev7 subunit of Polζ, which is a multifaceted HORMA (Hop1, Rev7, Mad2) protein with roles in TLS, DNA repair, and cell-cycle control, facilitates assembly of this complex by binding Rev1 and the catalytic subunit of Polζ, Rev3. Rev7 interacts with Rev3 by a mechanism conserved among HORMA proteins, whereby an open-to-closed transition locks the ligand underneath the "safety belt" loop. Dimerization of HORMA proteins promotes binding and release of this ligand, as exemplified by the Rev7 homolog, Mad2. Here, we investigate the dimerization of Rev7 when bound to the two Rev7-binding motifs (RBMs) in Rev3 by combining in vitro analyses of Rev7 structure and interactions with a functional assay in a Rev7 cell line. We demonstrate that Rev7 uses the conventional HORMA dimerization interface both to form a homodimer when tethered by the two RBMs in Rev3 and to heterodimerize with other HORMA domains, Mad2 and p31 Structurally, the Rev7 dimer can bind only one copy of Rev1, revealing an unexpected Rev1/Polζ architecture. In cells, mutation of the Rev7 dimer interface increases sensitivity to DNA damage. These results provide insights into the structure of the Rev1/Polζ TLS assembly and highlight the function of Rev7 homo- and heterodimerization. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDCY8 SASDCY8 |
|---|
-Related structure data
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #1623 |  Type: atomic / Radius of dummy atoms: 1.90 A Comment: residues at N- and C-termini that were missing in PDB were built in extended conformation Chi-square value: 9.75312841135  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: Human Rev7 monomer (R124A mutant) in complex with Rev3 peptide @ 8mg/mL Specimen concentration: 8 mg/ml / Entity id: 854 / 855 |
|---|---|
| Buffer | Name: 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol pH: 8.4 |
| Entity #854 | Name: hRev7-R124A / Type: protein Description: Mitotic spindle assembly checkpoint protein MAD2B Formula weight: 24.306 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q9UI95 Sequence: GMTTLTRQDL NFGQVVADVL CEFLEVAVHL ILYVREVYPV GIFQKRKKYN VPVQMSCHPE LNQYIQDTLH CVKPLLEKND VEKVVVVILD KEHRPVEKFV FEITQPPLLS ISSDSLLSHV EQLLAAFILK ISVCDAVLDH NPPGCTFTVL VHTREAATRN MEKIQVIKDF ...Sequence: GMTTLTRQDL NFGQVVADVL CEFLEVAVHL ILYVREVYPV GIFQKRKKYN VPVQMSCHPE LNQYIQDTLH CVKPLLEKND VEKVVVVILD KEHRPVEKFV FEITQPPLLS ISSDSLLSHV EQLLAAFILK ISVCDAVLDH NPPGCTFTVL VHTREAATRN MEKIQVIKDF PWILADEQDV HMHDPRLIPL KTMTSDILKM QLYVEERAHK GS |
| Entity #855 | Name: Rev3-RBM2 / Type: protein / Description: DNA polymerase zeta catalytic subunit / Formula weight: 3.273 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: O60673 Sequence: MEDKKIVIMP CKCAPSRQLV QVWLQAKE |
-Experimental information
| Beam | Instrument name: Cornell High Energy Synchrotron Source (CHESS) G1 City: Ithaca, NY / 国: USA  / Type of source: X-ray synchrotron / Wavelength: 0.1267 Å / Type of source: X-ray synchrotron / Wavelength: 0.1267 Å | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Finger Lakes CCD / Type: CCD | |||||||||||||||||||||||||||||||||||||||
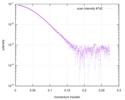
| Scan | Measurement date: May 14, 2016 / Storage temperature: 4 °C / Cell temperature: 4 °C / Exposure time: 1 sec. / Number of frames: 10 / Unit: 1/A /
| |||||||||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller