[English] 日本語
 Yorodumi
Yorodumi- SASDCQ9: Ks-Amt5-HK (+10uM ATP) (Ammonium transporter histidine kinase dom... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 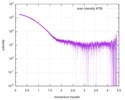 |
|---|---|
 Sample Sample | Ks-Amt5-HK (+10uM ATP)
|
| Function / homology |  Function and homology information Function and homology informationammonium homeostasis / ammonium channel activity / phosphorelay sensor kinase activity / plasma membrane Similarity search - Function |
| Biological species |  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Signaling ammonium across membranes through an ammonium sensor histidine kinase. Authors: Tobias Pflüger / Camila F Hernández / Philipp Lewe / Fabian Frank / Haydyn Mertens / Dmitri Svergun / Manfred W Baumstark / Vladimir Y Lunin / Mike S M Jetten / Susana L A Andrade /    Abstract: Sensing and uptake of external ammonium is essential for anaerobic ammonium-oxidizing (anammox) bacteria, and is typically the domain of the ubiquitous Amt/Rh ammonium transporters. Here, we report ...Sensing and uptake of external ammonium is essential for anaerobic ammonium-oxidizing (anammox) bacteria, and is typically the domain of the ubiquitous Amt/Rh ammonium transporters. Here, we report on the structure and function of an ammonium sensor/transducer from the anammox bacterium "Candidatus Kuenenia stuttgartiensis" that combines a membrane-integral ammonium transporter domain with a fused histidine kinase. It contains a high-affinity ammonium binding site not present in assimilatory Amt proteins. The levels of phosphorylated histidine in the kinase are coupled to the presence of ammonium, as conformational changes during signal recognition by the Amt module are transduced internally to modulate the kinase activity. The structural analysis of this ammonium sensor by X-ray crystallography and small-angle X-ray-scattering reveals a flexible, bipartite system that recruits a large uptake transporter as a sensory module and modulates its functionality to achieve a mechanistic coupling to a kinase domain in order to trigger downstream signaling events. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDCQ9 SASDCQ9 |
|---|
-Related structure data
| Related structure data |  6eu6C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Ks-Amt5-HK (+10uM ATP) / Specimen concentration: 1.00-5.00 |
|---|---|
| Buffer | Name: 20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol / pH: 8 |
| Entity #910 | Name: Ks-Amt5-HK / Type: protein / Description: Ammonium transporter histidine kinase domain / Formula weight: 30.031 / Num. of mol.: 1 / Source: Candidatus Kuenenia stuttgartiensis / References: UniProt: Q1Q357 Sequence: FIRITRLQDI NTILEKRVQE KTADLQMANV ALEKANRLKS EFLTTMSAEL RTPLNAIIGF AEVLRDEIAG SLSKDQKEYV TDIHSSGHHL LDMINNILDL SKIETGKMHL QYEEFCIEDA INDTLTIINA SANNKGISVH TNIQDNTPLL SADKTKFRQI LYNLLSNAVK ...Sequence: FIRITRLQDI NTILEKRVQE KTADLQMANV ALEKANRLKS EFLTTMSAEL RTPLNAIIGF AEVLRDEIAG SLSKDQKEYV TDIHSSGHHL LDMINNILDL SKIETGKMHL QYEEFCIEDA INDTLTIINA SANNKGISVH TNIQDNTPLL SADKTKFRQI LYNLLSNAVK FTPENGKITI NVFQKDNSLQ FEIVDTGIGI KPEDKEKLFE AFHQADASLT REYEGTGLGL HLTKRLVELH GGKIWAESTF GKGSTFFFIL PINPVNK |
-Experimental information
| Beam | Instrument name: PETRA III P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 2.7 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 2.7 mm | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||||||||||||||||||||
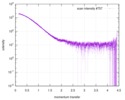
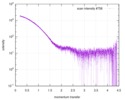
| Scan |
| ||||||||||||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller