+Search query
-Structure paper
| Title | Signaling ammonium across membranes through an ammonium sensor histidine kinase. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 9, Issue 1, Page 164, Year 2018 |
| Publish date | Jan 11, 2018 |
 Authors Authors | Tobias Pflüger / Camila F Hernández / Philipp Lewe / Fabian Frank / Haydyn Mertens / Dmitri Svergun / Manfred W Baumstark / Vladimir Y Lunin / Mike S M Jetten / Susana L A Andrade /    |
| PubMed Abstract | Sensing and uptake of external ammonium is essential for anaerobic ammonium-oxidizing (anammox) bacteria, and is typically the domain of the ubiquitous Amt/Rh ammonium transporters. Here, we report ...Sensing and uptake of external ammonium is essential for anaerobic ammonium-oxidizing (anammox) bacteria, and is typically the domain of the ubiquitous Amt/Rh ammonium transporters. Here, we report on the structure and function of an ammonium sensor/transducer from the anammox bacterium "Candidatus Kuenenia stuttgartiensis" that combines a membrane-integral ammonium transporter domain with a fused histidine kinase. It contains a high-affinity ammonium binding site not present in assimilatory Amt proteins. The levels of phosphorylated histidine in the kinase are coupled to the presence of ammonium, as conformational changes during signal recognition by the Amt module are transduced internally to modulate the kinase activity. The structural analysis of this ammonium sensor by X-ray crystallography and small-angle X-ray-scattering reveals a flexible, bipartite system that recruits a large uptake transporter as a sensory module and modulates its functionality to achieve a mechanistic coupling to a kinase domain in order to trigger downstream signaling events. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:29323112 / PubMed:29323112 /  PubMed Central PubMed Central |
| Methods | SAS (X-ray synchrotron) / X-ray diffraction |
| Resolution | 1.98 Å |
| Structure data |  SASDCM9: 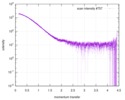 SASDCN9: Ks-Amt5-HK (apo) (Ammonium transporter histidine kinase domain, Ks-Amt5-HK) 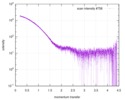 SASDCP9: Ks-Amt5-HK (+10uM APPCP) (Ammonium transporter histidine kinase domain, Ks-Amt5-HK) 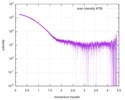 SASDCQ9: Ks-Amt5-HK (+10uM ATP) (Ammonium transporter histidine kinase domain, Ks-Amt5-HK)  SASDCS9: Ammonium transporter Ks-Amt5 (Batch) (ammonium sensor/transducer, Ks-Amt5)  PDB-6eu6: |
| Chemicals |  ChemComp-MES: 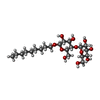 ChemComp-ZDM:  ChemComp-LMT:  ChemComp-NH4:  ChemComp-SO4:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Ammonium transporter / Signalling / histidine kinase |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 Candidatus kuenenia stuttgartiensis (bacteria)
Candidatus kuenenia stuttgartiensis (bacteria)