+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAN2 |
|---|---|
 Sample Sample | RNase in PBS
|
| Function / homology |  Function and homology information Function and homology informationpancreatic ribonuclease / ribonuclease A activity / RNA nuclease activity / nucleic acid binding / defense response to Gram-positive bacterium / hydrolase activity / extracellular region Similarity search - Function |
| Biological species |  |
 Contact author Contact author |
|
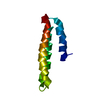
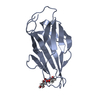
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDAN2 SASDAN2 |
|---|
-Related structure data
| Similar structure data |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #25 |  Type: dummy / Radius of dummy atoms: 1.90 A / Symmetry: P1 Comment: Gasbori run 10 times, damsel identified structure with lowest NSD Chi-square value: 2.4025  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #24 |  Type: atomic / Symmetry: P1 / Chi-square value: 1.493284  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: RNase in PBS / Ext coefficient: 6.9 / Contrast: 3.163 / Specific vol: 0.7279 / Purity method: gel filtration / Dry vol: 19917 / Sample MW: 16.46 kDa / Specimen concentration: 22.6 mg/ml / Concentration method: nanodrop |
|---|---|
| Buffer | Name: PBS / PK: 7 / pH: 7.4 |
| Entity #17 | Name: RNase / Type: protein / Description: Ribonuclease pancreatic / Formula weight: 16.46 / Num. of mol.: 1 / Source: Bos taurus / References: UniProt: P61823 Sequence: MALKSLVLLS LLVLVLLLVR VQPSLGKETA AAKFERQHMD SSTSAASSSN YCNQMMKSRN LTKDRCKPVN TFVHESLADV QAVCSQKNVA CKNGQTNCYQ SYSTMSITDC RETGSSKYPN CAYKTTQANK HIIVACEGNP YVPVHFDASV |
-Experimental information
| Beam | Instrument name: DORIS III EMBL X33 / City: Hamburg / 国: Germany  / Shape: 0.6 / Type of source: X-ray synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm / Shape: 0.6 / Type of source: X-ray synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M-W / Pixsize x: 0.172 mm | ||||||||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller