[English] 日本語
 Yorodumi
Yorodumi- PDB-8e50: Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8.0E+50 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA | ||||||
 Components Components | Glycerol-3-phosphate acyltransferase 1, mitochondrial | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / acyltransferase / LPA / monotopic / mitochondrial | ||||||
| Function / homology |  Function and homology information Function and homology informationglycerol-3-phosphate 1-O-acyltransferase / glycerol-3-phosphate O-acyltransferase activity / phosphatidylglycerol biosynthetic process / CDP-diacylglycerol biosynthetic process / Triglyceride biosynthesis / diacylglycerol biosynthetic process / glycerophospholipid metabolic process / negative regulation of activation-induced cell death of T cells / triglyceride biosynthetic process / phosphatidic acid biosynthetic process ...glycerol-3-phosphate 1-O-acyltransferase / glycerol-3-phosphate O-acyltransferase activity / phosphatidylglycerol biosynthetic process / CDP-diacylglycerol biosynthetic process / Triglyceride biosynthesis / diacylglycerol biosynthetic process / glycerophospholipid metabolic process / negative regulation of activation-induced cell death of T cells / triglyceride biosynthetic process / phosphatidic acid biosynthetic process / Synthesis of PA / activation-induced cell death of T cells / acyl-CoA metabolic process / phospholipid homeostasis / positive regulation of multicellular organism growth / activated T cell proliferation / RUNX1 regulates estrogen receptor mediated transcription / positive regulation of activated T cell proliferation / fatty acid homeostasis / response to glucose / regulation of cytokine production / Activation of gene expression by SREBF (SREBP) / fatty acid metabolic process / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / defense response to virus / Estrogen-dependent gene expression / mitochondrial outer membrane / mitochondrion / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.67 Å | ||||||
 Authors Authors | Wasilko, D.J. / Johnson, Z.L. / Ammirati, M. / Chang, J.S. / Han, S. / Wu, H. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis of the acyl-transfer mechanism of human GPAT1. Authors: Zachary Lee Johnson / Mark Ammirati / David Jonathan Wasilko / Jeanne S Chang / Stephen Noell / Timothy L Foley / Hyejin Yoon / Kathleen Smith / Shoh Asano / Katherine Hales / Min Wan / ...Authors: Zachary Lee Johnson / Mark Ammirati / David Jonathan Wasilko / Jeanne S Chang / Stephen Noell / Timothy L Foley / Hyejin Yoon / Kathleen Smith / Shoh Asano / Katherine Hales / Min Wan / Qingyi Yang / Mary A Piotrowski / Kathleen A Farley / Tamara Gilbert / Lisa M Aschenbrenner / Kimberly F Fennell / Jason K Dutra / Mary Xu / Chunyang Guo / Alison E Varghese / Justin Bellenger / Alandra Quinn / Christopher W Am Ende / Graham M West / Matthew C Griffor / Donald Bennett / Matthew Calabrese / Claire M Steppan / Seungil Han / Huixian Wu /  Abstract: Glycerol-3-phosphate acyltransferase (GPAT)1 is a mitochondrial outer membrane protein that catalyzes the first step of de novo glycerolipid biosynthesis. Hepatic expression of GPAT1 is linked to ...Glycerol-3-phosphate acyltransferase (GPAT)1 is a mitochondrial outer membrane protein that catalyzes the first step of de novo glycerolipid biosynthesis. Hepatic expression of GPAT1 is linked to liver fat accumulation and the severity of nonalcoholic fatty liver diseases. Here we present the cryo-EM structures of human GPAT1 in substrate analog-bound and product-bound states. The structures reveal an N-terminal acyltransferase domain that harbors important catalytic motifs and a tightly associated C-terminal domain that is critical for proper protein folding. Unexpectedly, GPAT1 has no transmembrane regions as previously proposed but instead associates with the membrane via an amphipathic surface patch and an N-terminal loop-helix region that contains a mitochondrial-targeting signal. Combined structural, computational and functional studies uncover a hydrophobic pathway within GPAT1 for lipid trafficking. The results presented herein lay a framework for rational inhibitor development for GPAT1. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8e50.cif.gz 8e50.cif.gz | 150.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8e50.ent.gz pdb8e50.ent.gz | 111.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8e50.json.gz 8e50.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e5/8e50 https://data.pdbj.org/pub/pdb/validation_reports/e5/8e50 ftp://data.pdbj.org/pub/pdb/validation_reports/e5/8e50 ftp://data.pdbj.org/pub/pdb/validation_reports/e5/8e50 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  27899MC  8e4yC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 86919.469 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GPAM, GPAT1, KIAA1560 / Production host: Homo sapiens (human) / Gene: GPAM, GPAT1, KIAA1560 / Production host:  References: UniProt: Q9HCL2, glycerol-3-phosphate 1-O-acyltransferase |
|---|---|
| #2: Chemical | ChemComp-COA / |
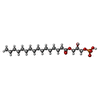
| #3: Chemical | ChemComp-NKO / ( |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: glycerol-3-phosphate acyltransferase 1 / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.086 MDa / Experimental value: NO | ||||||||||||||||||||||||||||||||
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||||
| Specimen | Conc.: 6.1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K / Details: Blot force -5, blot time 3 sec |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2200 nm / Nominal defocus min: 600 nm / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 9 sec. / Electron dose: 78 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 9153 |
| EM imaging optics | Energyfilter name: GIF Quantum LS / Energyfilter slit width: 20 eV |
| Image scans | Width: 3838 / Height: 3710 / Movie frames/image: 50 |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.8.0267 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Image processing | Details: collected in super resolution mode; gain normalized and binned by 2 during motion correction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1704123 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.67 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 76033 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 8E4Y Pdb chain-ID: A / Accession code: 8E4Y / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 3.67→98 Å / Cor.coef. Fo:Fc: 0.903 / SU B: 53.606 / SU ML: 0.691 / ESU R: 1.452 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 169.051 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Total: 5277 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj