[English] 日本語
 Yorodumi
Yorodumi- EMDB-9878: Cryo EM density map of Resveratrol-stabilized bioactive insulin o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9878 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM density map of Resveratrol-stabilized bioactive insulin oligomer | ||||||||||||
 Map data Map data | Resveratrol-stabilized oligomeric form of insulin which is fully biologically active | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Insulin fibrillation / Natural polyphenols / Anti-Amyloid activity / Insulin hexamer / Bioavailability / HORMONE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of glycogen catabolic process / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / regulation of protein secretion / Insulin processing / positive regulation of peptide hormone secretion / positive regulation of respiratory burst ...negative regulation of glycogen catabolic process / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / regulation of protein secretion / Insulin processing / positive regulation of peptide hormone secretion / positive regulation of respiratory burst / negative regulation of acute inflammatory response / Regulation of gene expression in beta cells / alpha-beta T cell activation / positive regulation of dendritic spine maintenance / Synthesis, secretion, and deacylation of Ghrelin / activation of protein kinase B activity / negative regulation of protein secretion / negative regulation of gluconeogenesis / positive regulation of glycogen biosynthetic process / fatty acid homeostasis / Signal attenuation / positive regulation of insulin receptor signaling pathway / negative regulation of respiratory burst involved in inflammatory response / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / negative regulation of lipid catabolic process / positive regulation of lipid biosynthetic process / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / regulation of protein localization to plasma membrane / nitric oxide-cGMP-mediated signaling / transport vesicle / COPI-mediated anterograde transport / positive regulation of nitric-oxide synthase activity / Insulin receptor recycling / negative regulation of reactive oxygen species biosynthetic process / positive regulation of brown fat cell differentiation / insulin-like growth factor receptor binding / NPAS4 regulates expression of target genes / neuron projection maintenance / endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of mitotic nuclear division / Insulin receptor signalling cascade / positive regulation of glycolytic process / positive regulation of cytokine production / endosome lumen / positive regulation of long-term synaptic potentiation / acute-phase response / positive regulation of protein secretion / positive regulation of D-glucose import across plasma membrane / insulin receptor binding / positive regulation of cell differentiation / Regulation of insulin secretion / wound healing / positive regulation of neuron projection development / hormone activity / regulation of synaptic plasticity / negative regulation of protein catabolic process / positive regulation of protein localization to nucleus / Golgi lumen / vasodilation / cognition / glucose metabolic process / insulin receptor signaling pathway / cell-cell signaling / glucose homeostasis / regulation of protein localization / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / positive regulation of cell growth / protease binding / secretory granule lumen / positive regulation of canonical NF-kappaB signal transduction / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of MAPK cascade / positive regulation of cell migration / G protein-coupled receptor signaling pathway / endoplasmic reticulum lumen / Amyloid fiber formation / Golgi membrane / negative regulation of gene expression / positive regulation of cell population proliferation / positive regulation of gene expression / regulation of DNA-templated transcription / extracellular space / extracellular region / identical protein binding Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
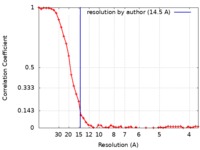
| Method | single particle reconstruction / cryo EM / Resolution: 14.5 Å | ||||||||||||
 Authors Authors | Sengupta J / Pathak BK | ||||||||||||
| Funding support |  India, 3 items India, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Comput Aided Mol Des / Year: 2020 Journal: J Comput Aided Mol Des / Year: 2020Title: Resveratrol as a nontoxic excipient stabilizes insulin in a bioactive hexameric form. Authors: Bani Kumar Pathak / Debajyoti Das / Sayan Bhakta / Partha Chakrabarti / Jayati Sengupta /  Abstract: Insulin aggregation is the leading cause of considerable reduction in the amount of active drug molecules in liquid formulations manufactured for diabetes management. Phenolic compounds, such as ...Insulin aggregation is the leading cause of considerable reduction in the amount of active drug molecules in liquid formulations manufactured for diabetes management. Phenolic compounds, such as phenol and m-cresol, are routinely used to stabilize insulin in a hexameric form during its commercial preparation. However, long term usage of commercial insulin results in various adverse secondary responses, for which toxicity of the phenolic excipients is primarily responsible. In this study we aimed to find out a nontoxic insulin stabilizer. To that end, we have selected resveratrol, a natural polyphenol, as a prospective nontoxic insulin stabilizer because of its structural similarity with commercially used phenolic compounds. Atomic force microscopy visualization of resveratrol-treated human insulin revealed that resveratrol has a unique ability to arrest hINS in a soluble oligomeric form having discrete spherical morphology. Most importantly, resveratrol-treated insulin is nontoxic for HepG2 cells and it effectively maintains low blood glucose in a mouse model. Cryo-electron microscopy revealed 3D morphology of resveratrol-stabilized insulin that strikingly resembles crystal structures of insulin hexamer formulated with m-cresol. Significantly, we found that, in a condition inductive to amyloid fibrillation at physiological pH, resveratrol is capable of stabilizing insulin more efficiently than m-cresol. Thus, this study describes resveratrol as an effective nontoxic natural molecule that can be used for stabilizing insulin in a bioactive oligomeric form during its commercial formulation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9878.map.gz emd_9878.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9878-v30.xml emd-9878-v30.xml emd-9878.xml emd-9878.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9878_fsc.xml emd_9878_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_9878.png emd_9878.png | 439.7 KB | ||
| Filedesc metadata |  emd-9878.cif.gz emd-9878.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9878 http://ftp.pdbj.org/pub/emdb/structures/EMD-9878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9878 | HTTPS FTP |
-Validation report
| Summary document |  emd_9878_validation.pdf.gz emd_9878_validation.pdf.gz | 410.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9878_full_validation.pdf.gz emd_9878_full_validation.pdf.gz | 410.3 KB | Display | |
| Data in XML |  emd_9878_validation.xml.gz emd_9878_validation.xml.gz | 8 KB | Display | |
| Data in CIF |  emd_9878_validation.cif.gz emd_9878_validation.cif.gz | 10.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9878 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9878 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9878 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9878 | HTTPS FTP |
-Related structure data
| Related structure data |  6jr3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9878.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9878.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Resveratrol-stabilized oligomeric form of insulin which is fully biologically active | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.89 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Insulin oligomer and Resveratrol complex
| Entire | Name: Insulin oligomer and Resveratrol complex |
|---|---|
| Components |
|
-Supramolecule #1: Insulin oligomer and Resveratrol complex
| Supramolecule | Name: Insulin oligomer and Resveratrol complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Insulin A chain
| Macromolecule | Name: Insulin A chain / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.383698 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIVEQCCTSI CSLYQLENYC N UniProtKB: Insulin |
-Macromolecule #2: Insulin B chain
| Macromolecule | Name: Insulin B chain / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.433953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FVNQHLCGSH LVEALYLVCG ERGFFYTPKT UniProtKB: Insulin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)