+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8015 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Lysenin Pore | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pore forming protein / aerolysin / toxin | |||||||||
| Function / homology | other organism cell membrane / monoatomic ion transport / toxin activity / killing of cells of another organism / defense response to bacterium / extracellular region / membrane / Lysenin Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Savva CG / Bokori-Brown M | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Authors: Monika Bokori-Brown / Thomas G Martin / Claire E Naylor / Ajit K Basak / Richard W Titball / Christos G Savva /  Abstract: Lysenin from the coelomic fluid of the earthworm Eisenia fetida belongs to the aerolysin family of small β-pore-forming toxins (β-PFTs), some members of which are pathogenic to humans and animals. ...Lysenin from the coelomic fluid of the earthworm Eisenia fetida belongs to the aerolysin family of small β-pore-forming toxins (β-PFTs), some members of which are pathogenic to humans and animals. Despite efforts, a high-resolution structure of a channel for this family of proteins has been elusive and therefore the mechanism of activation and membrane insertion remains unclear. Here we determine the pore structure of lysenin by single particle cryo-EM, to 3.1 Å resolution. The nonameric assembly reveals a long β-barrel channel spanning the length of the complex that, unexpectedly, includes the two pre-insertion strands flanking the hypothetical membrane-insertion loop. Examination of other members of the aerolysin family reveals high structural preservation in this region, indicating that the membrane-insertion pathway in this family is conserved. For some toxins, proteolytic activation and pro-peptide removal will facilitate unfolding of the pre-insertion strands, allowing them to form the β-barrel of the channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8015.map.gz emd_8015.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8015-v30.xml emd-8015-v30.xml emd-8015.xml emd-8015.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8015_1.png emd_8015_1.png emd_8015_2.png emd_8015_2.png | 202.3 KB 252.9 KB | ||
| Filedesc metadata |  emd-8015.cif.gz emd-8015.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8015 http://ftp.pdbj.org/pub/emdb/structures/EMD-8015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8015 | HTTPS FTP |
-Related structure data
| Related structure data |  5gaqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8015.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8015.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.43 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
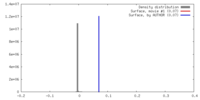
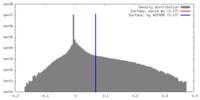
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Membrane-inserted form of the nonameric pore forming protein Lysenin
| Entire | Name: Membrane-inserted form of the nonameric pore forming protein Lysenin |
|---|---|
| Components |
|
-Supramolecule #1: Membrane-inserted form of the nonameric pore forming protein Lysenin
| Supramolecule | Name: Membrane-inserted form of the nonameric pore forming protein Lysenin type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 315 KDa |
-Macromolecule #1: Lysenin
| Macromolecule | Name: Lysenin / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.977246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAKAAEGYE QIEVDVVAVW KEGYVYENRG STSVDQKITI TKGMKNVNSE TRTVTATHSI GSTISTGDAF EIGSVEVSYS HSHEESQVS MTETEVYESK VIEHTITIPP TSKFTRWQLN ADVGGADIEY MYLIDEVTPI GGTQSIPQVI TSRAKIIVGR Q IILGKTEI ...String: MSAKAAEGYE QIEVDVVAVW KEGYVYENRG STSVDQKITI TKGMKNVNSE TRTVTATHSI GSTISTGDAF EIGSVEVSYS HSHEESQVS MTETEVYESK VIEHTITIPP TSKFTRWQLN ADVGGADIEY MYLIDEVTPI GGTQSIPQVI TSRAKIIVGR Q IILGKTEI RIKHAERKEY MTVVSRKSWP AATLGHSKLF KFVLYEDWGG FRIKTLNTMY SGYEYAYSSD QGGIYFDQGT DN PKQRWAI NKSLPLRHGD VVTFMNKYFT RSGLCYDDGP ATNVYCLDKR EDKWILEVVG LVPRGSGHHH HHH UniProtKB: Lysenin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS Details: Grids were glow-discharged prior to deposition of Graphene-oxide.No further treatment was performed after graphene-oxide deposition. | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 85.0 K / Max: 85.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | Direct alignments: Beam tilt pivot points, Beam shift, Comma Free. C2 aperture centering, C2 lens astigmatism correction. Objective aperture centering and objective lens astigmatism correction. Energy filter Tuning and occasional ZLP centering. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 0-20 / Number grids imaged: 1 / Number real images: 280 / Average exposure time: 18.0 sec. / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 4.7 µm / Calibrated defocus min: 0.7000000000000001 µm / Calibrated magnification: 34965 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 150-297 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Initial docking of C-terminal region of lysenin monomer to EM map was performed using UCSF Chimera followed by model N-terminal extension in COOT. |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-5gaq: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)