[English] 日本語
 Yorodumi
Yorodumi- EMDB-7453: Structure of the HIV-Nef dileucine mutant bound AP-1:Arf1 dimer m... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7453 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the HIV-Nef dileucine mutant bound AP-1:Arf1 dimer monomeric subunit | |||||||||
 Map data Map data | AP-1:Arf1:Tetherin-Nef dileucine mutant dimer monomeric subunit | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of plasmacytoid dendritic cell cytokine production / negative regulation of intracellular transport of viral material / response to interferon-beta / basolateral protein secretion / Lysosome Vesicle Biogenesis / endosome to melanosome transport / AP-1 adaptor complex / mitotic cleavage furrow ingression / trans-Golgi Network Vesicle Budding / protein trimerization ...negative regulation of plasmacytoid dendritic cell cytokine production / negative regulation of intracellular transport of viral material / response to interferon-beta / basolateral protein secretion / Lysosome Vesicle Biogenesis / endosome to melanosome transport / AP-1 adaptor complex / mitotic cleavage furrow ingression / trans-Golgi Network Vesicle Budding / protein trimerization / response to interferon-alpha / negative regulation of glycoprotein biosynthetic process / platelet dense granule organization / Glycosphingolipid transport / metalloendopeptidase inhibitor activity / symbiont-mediated suppression of host antigen processing and presentation of peptide antigen via MHC class I / melanosome assembly / regulation of receptor internalization / Intra-Golgi traffic / symbiont-mediated suppression of host antigen processing and presentation of peptide antigen via MHC class II / regulation of Arp2/3 complex-mediated actin nucleation / Golgi Associated Vesicle Biogenesis / Synthesis of PIPs at the Golgi membrane / symbiont-mediated suppression of host autophagy / clathrin-cargo adaptor activity / symbiont-mediated suppression of host apoptosis / positive regulation of leukocyte proliferation / MHC class II antigen presentation / thioesterase binding / Nef Mediated CD4 Down-regulation / CD4 receptor binding / dendritic spine organization / long-term synaptic depression / determination of left/right symmetry / clathrin-coated vesicle / COPI-dependent Golgi-to-ER retrograde traffic / azurophil granule membrane / clathrin binding / negative regulation of viral genome replication / Lysosome Vesicle Biogenesis / Golgi Associated Vesicle Biogenesis / Synthesis of PIPs at the plasma membrane / cell leading edge / MHC class I protein binding / B cell activation / host cell Golgi membrane / intracellular copper ion homeostasis / response to type II interferon / protein targeting / side of membrane / COPI-mediated anterograde transport / vesicle-mediated transport / viral life cycle / multivesicular body / regulation of calcium-mediated signaling / clathrin-coated pit / Neutrophil degranulation / MHC class II antigen presentation / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / negative regulation of cell migration / cytoplasmic vesicle membrane / Nef mediated downregulation of MHC class I complex cell surface expression / sarcomere / trans-Golgi network membrane / small monomeric GTPase / regulation of actin cytoskeleton organization / intracellular protein transport / kidney development / trans-Golgi network / negative regulation of cell growth / cellular response to virus / SH3 domain binding / virion component / response to virus / synaptic vesicle / SARS-CoV-1 activates/modulates innate immune responses / Interferon alpha/beta signaling / presynapse / heart development / ATPase binding / defense response to virus / early endosome / positive regulation of canonical NF-kappaB signal transduction / neuron projection / postsynaptic density / symbiont-mediated suppression of host innate immune response / apical plasma membrane / membrane raft / protein domain specific binding / Golgi membrane / signaling receptor binding / innate immune response / lysosomal membrane / focal adhesion / intracellular membrane-bounded organelle / GTPase activity / synapse / Neutrophil degranulation / protein kinase binding / GTP binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.27 Å | |||||||||
 Authors Authors | Buffalo CZ / Morris KL / Hurley JH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation. Authors: Kyle L Morris / Cosmo Z Buffalo / Christina M Stürzel / Elena Heusinger / Frank Kirchhoff / Xuefeng Ren / James H Hurley /   Abstract: The HIV accessory protein Nef counteracts immune defenses by subverting coated vesicle pathways. The 3.7 Å cryo-EM structure of a closed trimer of the clathrin adaptor AP-1, the small GTPase Arf1, ...The HIV accessory protein Nef counteracts immune defenses by subverting coated vesicle pathways. The 3.7 Å cryo-EM structure of a closed trimer of the clathrin adaptor AP-1, the small GTPase Arf1, HIV-1 Nef, and the cytosolic tail of the restriction factor tetherin suggested a mechanism for inactivating tetherin by Golgi retention. The 4.3 Å structure of a mutant Nef-induced dimer of AP-1 showed how the closed trimer is regulated by the dileucine loop of Nef. HDX-MS and mutational analysis were used to show how cargo dynamics leads to alternative Arf1 trimerization, directing Nef targets to be either retained at the trans-Golgi or sorted to lysosomes. Phosphorylation of the NL4-3 M-Nef was shown to regulate AP-1 trimerization, explaining how O-Nefs lacking this phosphosite counteract tetherin but most M-Nefs do not. These observations show how the higher-order organization of a vesicular coat can be allosterically modulated to direct cargoes to distinct fates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7453.map.gz emd_7453.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7453-v30.xml emd-7453-v30.xml emd-7453.xml emd-7453.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7453.png emd_7453.png | 643.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7453 http://ftp.pdbj.org/pub/emdb/structures/EMD-7453 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7453 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7453 | HTTPS FTP |
-Related structure data
| Related structure data |  6d83MC  7454C  7455C  7456C  7457C  7458C  7563C  6cm9C  6criC  6d84C  6dffC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7453.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7453.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AP-1:Arf1:Tetherin-Nef dileucine mutant dimer monomeric subunit | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
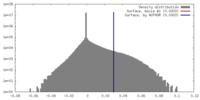
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dimer of AP-1:Arf1:Tetherin-Nef dileucine mutant
| Entire | Name: Dimer of AP-1:Arf1:Tetherin-Nef dileucine mutant |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of AP-1:Arf1:Tetherin-Nef dileucine mutant
| Supramolecule | Name: Dimer of AP-1:Arf1:Tetherin-Nef dileucine mutant / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 230 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris at pH 8.0, 200 mM NaCl, 5 mM MgCl2, and 0.5 mM TCEP |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK IV |
| Details | AP-1:Arf1:Tetherin-Nef dileucine (LL164-165AA) mutant |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3712 pixel / Digitization - Dimensions - Height: 3840 pixel / Digitization - Frames/image: 3-28 / Number grids imaged: 1 / Number real images: 1989 / Average exposure time: 5.6 sec. / Average electron dose: 39.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 46849 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.6 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)