+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6398 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of a Propionibacterium acnes phage | |||||||||
 Map data Map data | Reconstruction of Propionibacterium acnes phage | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | acne / bacteriophage / cryoEM / HK97-like | |||||||||
| Function / homology | Gp6 Function and homology information Function and homology information | |||||||||
| Biological species |  Propionibacterium phage PA6 (virus) Propionibacterium phage PA6 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Chiou J / Zhang X / Marinelli LJ / Modlin RL / Zhou ZH | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Capsid Structure of the Propionibacterium acnes Bacteriophage ATCC_Clear Authors: Chiou J / Zhang X / Marinelli LJ / Modlin RL / Zhou ZH | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6398.map.gz emd_6398.map.gz | 466 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6398-v30.xml emd-6398-v30.xml emd-6398.xml emd-6398.xml | 8.9 KB 8.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6398.jpg emd_6398.jpg | 205.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6398 http://ftp.pdbj.org/pub/emdb/structures/EMD-6398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6398 | HTTPS FTP |
-Related structure data
| Related structure data |  3jb5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6398.map.gz / Format: CCP4 / Size: 523.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6398.map.gz / Format: CCP4 / Size: 523.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Propionibacterium acnes phage | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
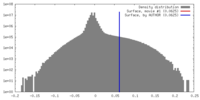
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Propionibacterium acnes bacteriophage ATCC_Clear capsid structure
| Entire | Name: Propionibacterium acnes bacteriophage ATCC_Clear capsid structure |
|---|---|
| Components |
|
-Supramolecule #1000: Propionibacterium acnes bacteriophage ATCC_Clear capsid structure
| Supramolecule | Name: Propionibacterium acnes bacteriophage ATCC_Clear capsid structure type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 33 MDa / Method: SDS-PAGE/MS |
-Supramolecule #1: Propionibacterium phage PA6
| Supramolecule | Name: Propionibacterium phage PA6 / type: virus / ID: 1 / Name.synonym: Propionibacterium acnes bacteriophage / Details: Mature virions, DNA inside masked / NCBI-ID: 376758 / Sci species name: Propionibacterium phage PA6 / Sci species strain: ATCC 29399B_C / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: Propionibacterium acnes bacteriophage |
|---|---|
| Host (natural) | Organism:  Propionibacterium acnes (bacteria) / Strain: 6919 / synonym: BACTERIA(EUBACTERIA) Propionibacterium acnes (bacteria) / Strain: 6919 / synonym: BACTERIA(EUBACTERIA) |
| Virus shell | Shell ID: 1 / Diameter: 660 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: Purified sample was applied to a Quantifoil grid |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 15 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | May 16, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 3168 / Average electron dose: 25 e/Å2 Details: Every image is the average of twelve frames recorded by the direct electron detector. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 38462 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.27 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 38462 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected using an in-house selection procedure. |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: OTHER / Software - Name: FREALIGN / Number images used: 27504 |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)