[English] 日本語
 Yorodumi
Yorodumi- EMDB-60917: Identification, structure and agonist design of an androgen membr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Identification, structure and agonist design of an androgen membrane receptor. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / agonist / GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of skeletal muscle contraction / G protein-coupled receptor activity / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels ...regulation of skeletal muscle contraction / G protein-coupled receptor activity / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / nuclear speck / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Ping YQ / Yang Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structural basis for the tethered peptide activation of adhesion GPCRs. Authors: Yu-Qi Ping / Peng Xiao / Fan Yang / Ru-Jia Zhao / Sheng-Chao Guo / Xu Yan / Xiang Wu / Chao Zhang / Yan Lu / Fenghui Zhao / Fulai Zhou / Yue-Tong Xi / Wanchao Yin / Feng-Zhen Liu / Dong-Fang ...Authors: Yu-Qi Ping / Peng Xiao / Fan Yang / Ru-Jia Zhao / Sheng-Chao Guo / Xu Yan / Xiang Wu / Chao Zhang / Yan Lu / Fenghui Zhao / Fulai Zhou / Yue-Tong Xi / Wanchao Yin / Feng-Zhen Liu / Dong-Fang He / Dao-Lai Zhang / Zhong-Liang Zhu / Yi Jiang / Lutao Du / Shi-Qing Feng / Torsten Schöneberg / Ines Liebscher / H Eric Xu / Jin-Peng Sun /   Abstract: Adhesion G-protein-coupled receptors (aGPCRs) are important for organogenesis, neurodevelopment, reproduction and other processes. Many aGPCRs are activated by a conserved internal (tethered) agonist ...Adhesion G-protein-coupled receptors (aGPCRs) are important for organogenesis, neurodevelopment, reproduction and other processes. Many aGPCRs are activated by a conserved internal (tethered) agonist sequence known as the Stachel sequence. Here, we report the cryogenic electron microscopy (cryo-EM) structures of two aGPCRs in complex with G: GPR133 and GPR114. The structures indicate that the Stachel sequences of both receptors assume an α-helical-bulge-β-sheet structure and insert into a binding site formed by the transmembrane domain (TMD). A hydrophobic interaction motif (HIM) within the Stachel sequence mediates most of the intramolecular interactions with the TMD. Combined with the cryo-EM structures, biochemical characterization of the HIM motif provides insight into the cross-reactivity and selectivity of the Stachel sequences. Two interconnected mechanisms, the sensing of Stachel sequences by the conserved 'toggle switch' W and the constitution of a hydrogen-bond network formed by Q/Y and the P/VφφG motif (φ indicates a hydrophobic residue), are important in Stachel sequence-mediated receptor activation and G coupling. Notably, this network stabilizes kink formation in TM helices 6 and 7 (TM6 and TM7, respectively). A common G-binding interface is observed between the two aGPCRs, and GPR114 has an extended TM7 that forms unique interactions with G. Our structures reveal the detailed mechanisms of aGPCR activation by Stachel sequences and their G coupling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60917.map.gz emd_60917.map.gz | 52.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60917-v30.xml emd-60917-v30.xml emd-60917.xml emd-60917.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60917.png emd_60917.png | 94.6 KB | ||
| Masks |  emd_60917_msk_1.map emd_60917_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-60917.cif.gz emd-60917.cif.gz | 7.3 KB | ||
| Others |  emd_60917_half_map_1.map.gz emd_60917_half_map_1.map.gz emd_60917_half_map_2.map.gz emd_60917_half_map_2.map.gz | 55.3 MB 55.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60917 http://ftp.pdbj.org/pub/emdb/structures/EMD-60917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60917 | HTTPS FTP |
-Related structure data
| Related structure data |  9iv1MC  8x9sC  8x9tC  8x9uC  9iv2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60917.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60917.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_60917_msk_1.map emd_60917_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
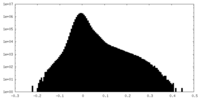
| Density Histograms |
-Half map: #2
| File | emd_60917_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
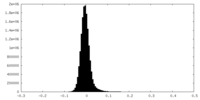
| Density Histograms |
-Half map: #1
| File | emd_60917_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of GPR133 with agonist and G protein trime
| Entire | Name: Complex of GPR133 with agonist and G protein trime |
|---|---|
| Components |
|
-Supramolecule #1: Complex of GPR133 with agonist and G protein trime
| Supramolecule | Name: Complex of GPR133 with agonist and G protein trime / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 28.781988 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SAGGGGSGGG GSGGGGSSDI VMTQATSSVP VTPGESVSIS C RSSKSLLH ...String: MVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SAGGGGSGGG GSGGGGSSDI VMTQATSSVP VTPGESVSIS C RSSKSLLH SNGNTYLYWF LQRPGQSPQL LIYRMSNLAS GVPDRFSGSG SGTAFTLTIS RLEAEDVGVY YCMQHLEYPL TF GAGTKLE LLEENLYFQG ASHHHHHHHH |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.784301 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSLLQSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT ...String: GSLLQSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT TCALWDIETG QQTTTFTGHT GDVMSLSLAP DTRLFVSGAC DASAKLWDVR EGMCRQTFTG HESDINAICF FP NGNAFAT GSDDATCRLF DLRADQELMT YSHDNIICGI TSVSFSKSGR LLLAGYDDFN CNVWDALKAD RAGVLAGHDN RVS CLGVTD DGMAVATGSW DSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Adhesion G-protein coupled receptor D1
| Macromolecule | Name: Adhesion G-protein coupled receptor D1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.24407 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HQVALSSISY VGCSLSVLCL VATLVTFAVL SSVSTIRNQR YHIHANLSFA VLVAQVLLLI SFRLEPGTTP CQVMAVLLHY FFLSAFAWM LVEGLHLYSM VIKVFGSEDS KHRYYYGMGW GFPLLICIIS LSFAMDSYGT SNNCWLSLAS GAIWAFVAPA L FVIVVNIG ...String: HQVALSSISY VGCSLSVLCL VATLVTFAVL SSVSTIRNQR YHIHANLSFA VLVAQVLLLI SFRLEPGTTP CQVMAVLLHY FFLSAFAWM LVEGLHLYSM VIKVFGSEDS KHRYYYGMGW GFPLLICIIS LSFAMDSYGT SNNCWLSLAS GAIWAFVAPA L FVIVVNIG ILIAVTRVIS QISADNYKIH GDPSAFKLTA KAVAVLLPIL GTSWVFGVLA VNGCAVVFQY MFATLNSLQG LF IFLFHCL LNSEVRAAFK HKTKVWSLTS SSARTSNAKP FHSDLMNGTR PGMASTKLSP WDKSSHSAHR VDLSAV UniProtKB: Adhesion G-protein coupled receptor D1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.879465 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RATHRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RATHRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTSGIFETK FQVDKVNFHM FDVGAQRDER RKWIQCFNDV TAIIFVVDSS DYNRLQEALN DF KSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED YFPEFARYTT PEDATPEPGE DPRVTRAKYF IRDEFLRIST ASG DGRHYC YPHFTCSVDT ENARRIFNDC RDIIQRMHLR QYELL |
-Macromolecule #6: 5-ALPHA-DIHYDROTESTOSTERONE
| Macromolecule | Name: 5-ALPHA-DIHYDROTESTOSTERONE / type: ligand / ID: 6 / Number of copies: 1 / Formula: DHT |
|---|---|
| Molecular weight | Theoretical: 290.44 Da |
| Chemical component information | 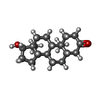 ChemComp-DHT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)