[English] 日本語
 Yorodumi
Yorodumi- EMDB-45803: Structural basis of BAK sequestration by MCL-1 and consequences f... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis of BAK sequestration by MCL-1 and consequences for apoptosis initiation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Anti-apoptosis / Mitochondrial poration / BCL-2 family / Cell fate / APOPTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationActivation and oligomerization of BAK protein / BH domain binding / B cell negative selection / BAK complex / negative regulation of endoplasmic reticulum calcium ion concentration / response to fungus / response to mycotoxin / Release of apoptotic factors from the mitochondria / limb morphogenesis / apoptotic process involved in blood vessel morphogenesis ...Activation and oligomerization of BAK protein / BH domain binding / B cell negative selection / BAK complex / negative regulation of endoplasmic reticulum calcium ion concentration / response to fungus / response to mycotoxin / Release of apoptotic factors from the mitochondria / limb morphogenesis / apoptotic process involved in blood vessel morphogenesis / post-embryonic camera-type eye morphogenesis / endocrine pancreas development / establishment or maintenance of transmembrane electrochemical gradient / B cell apoptotic process / negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / endoplasmic reticulum calcium ion homeostasis / positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / regulation of mitochondrial membrane permeability / calcium ion transport into cytosol / fibroblast apoptotic process / response to UV-C / mitochondrial fusion / Bcl-2 family protein complex / myeloid cell homeostasis / porin activity / thymocyte apoptotic process / pore complex / negative regulation of release of cytochrome c from mitochondria / positive regulation of IRE1-mediated unfolded protein response / positive regulation of release of cytochrome c from mitochondria / vagina development / B cell homeostasis / positive regulation of calcium ion transport into cytosol / positive regulation of proteolysis / intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress / blood vessel remodeling / animal organ regeneration / cellular response to unfolded protein / Pyroptosis / heat shock protein binding / extrinsic apoptotic signaling pathway in absence of ligand / intrinsic apoptotic signaling pathway / release of cytochrome c from mitochondria / epithelial cell proliferation / response to gamma radiation / regulation of mitochondrial membrane potential / apoptotic signaling pathway / response to hydrogen peroxide / establishment of localization in cell / positive regulation of protein-containing complex assembly / cellular response to mechanical stimulus / intrinsic apoptotic signaling pathway in response to DNA damage / cellular response to UV / protein-folding chaperone binding / channel activity / response to ethanol / transmembrane transporter binding / mitochondrial outer membrane / regulation of cell cycle / positive regulation of apoptotic process / response to xenobiotic stimulus / protein heterodimerization activity / negative regulation of cell population proliferation / negative regulation of gene expression / apoptotic process / protein-containing complex binding / endoplasmic reticulum / protein homodimerization activity / mitochondrion / metal ion binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.34 Å | |||||||||
 Authors Authors | Uchikawa E / Myasnikov A / Dey R / Moldoveanu T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2025 Journal: Mol Cell / Year: 2025Title: Structural basis of BAK sequestration by MCL-1 in apoptosis. Authors: Shagun Srivastava / Giridhar Sekar / Adedolapo Ojoawo / Anup Aggarwal / Elisabeth Ferreira / Emiko Uchikawa / Meek Yang / Christy R Grace / Raja Dey / Yi-Lun Lin / Cristina D Guibao / ...Authors: Shagun Srivastava / Giridhar Sekar / Adedolapo Ojoawo / Anup Aggarwal / Elisabeth Ferreira / Emiko Uchikawa / Meek Yang / Christy R Grace / Raja Dey / Yi-Lun Lin / Cristina D Guibao / Seetharaman Jayaraman / Somnath Mukherjee / Anthony A Kossiakoff / Bin Dong / Alexander Myasnikov / Tudor Moldoveanu /   Abstract: Apoptosis controls cell fate, ensuring tissue homeostasis and promoting disease when dysregulated. The rate-limiting step in apoptosis is mitochondrial poration by the effector B cell lymphoma 2 (BCL- ...Apoptosis controls cell fate, ensuring tissue homeostasis and promoting disease when dysregulated. The rate-limiting step in apoptosis is mitochondrial poration by the effector B cell lymphoma 2 (BCL-2) family proteins BAK and BAX, which are activated by initiator BCL-2 homology 3 (BH3)-only proteins (e.g., BIM) and inhibited by guardian BCL-2 family proteins (e.g., MCL-1). We integrated structural, biochemical, and pharmacological approaches to characterize the human prosurvival MCL-1:BAK complex assembled from their BCL-2 globular core domains. We reveal a canonical interaction with BAK BH3 bound to the hydrophobic groove of MCL-1 and disordered and highly dynamic BAK regions outside the complex interface. We predict similar conformations of activated effectors in complex with other guardians or effectors. The MCL-1:BAK complex is a major cancer drug target. We show that MCL-1 inhibitors are inefficient in neutralizing the MCL-1:BAK complex, requiring high doses to initiate apoptosis. Our study underscores the need to design superior clinical candidate MCL-1 inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45803.map.gz emd_45803.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45803-v30.xml emd-45803-v30.xml emd-45803.xml emd-45803.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_45803.png emd_45803.png | 81.5 KB | ||
| Filedesc metadata |  emd-45803.cif.gz emd-45803.cif.gz | 6.4 KB | ||
| Others |  emd_45803_half_map_1.map.gz emd_45803_half_map_1.map.gz emd_45803_half_map_2.map.gz emd_45803_half_map_2.map.gz | 638.2 KB 638 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45803 http://ftp.pdbj.org/pub/emdb/structures/EMD-45803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45803 | HTTPS FTP |
-Validation report
| Summary document |  emd_45803_validation.pdf.gz emd_45803_validation.pdf.gz | 409 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45803_full_validation.pdf.gz emd_45803_full_validation.pdf.gz | 408.5 KB | Display | |
| Data in XML |  emd_45803_validation.xml.gz emd_45803_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_45803_validation.cif.gz emd_45803_validation.cif.gz | 14.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45803 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45803 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45803 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45803 | HTTPS FTP |
-Related structure data
| Related structure data |  9cphMC  9cpeC  9cpfC  9cpnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45803.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45803.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.452 Å | ||||||||||||||||||||||||||||||||||||
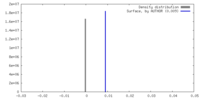
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_45803_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
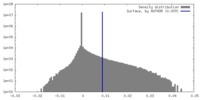
| Density Histograms |
-Half map: #1
| File | emd_45803_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
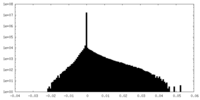
| Density Histograms |
- Sample components
Sample components
-Entire : hetero-tetrameric Complex of Sab11M heavy and long chain, MBP_MCL...
| Entire | Name: hetero-tetrameric Complex of Sab11M heavy and long chain, MBP_MCL1 fusion protein, and BAK-BH3 |
|---|---|
| Components |
|
-Supramolecule #1: hetero-tetrameric Complex of Sab11M heavy and long chain, MBP_MCL...
| Supramolecule | Name: hetero-tetrameric Complex of Sab11M heavy and long chain, MBP_MCL1 fusion protein, and BAK-BH3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Induced myeloid leukemia cell differentiation protein Mcl-1
| Macromolecule | Name: Induced myeloid leukemia cell differentiation protein Mcl-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.152789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KIEEGKLVIW INGDKGYNGL AEVGKKFEKD TGIKVTVEHP DKLEEKFPQV AATGDGPDII FWAHDRFGGY AQSGLLAEIT PDKAFQDKL YPFTWDAVRY NGKLIAYPIA VEALSLIYNK DLLPNPPKTW EEIPALDKEL KAKGKSALMF NLQEPYFTWP L IAADGGYA ...String: KIEEGKLVIW INGDKGYNGL AEVGKKFEKD TGIKVTVEHP DKLEEKFPQV AATGDGPDII FWAHDRFGGY AQSGLLAEIT PDKAFQDKL YPFTWDAVRY NGKLIAYPIA VEALSLIYNK DLLPNPPKTW EEIPALDKEL KAKGKSALMF NLQEPYFTWP L IAADGGYA FKYENGKYDI KDVGVDNAGA KAGLTFLVDL IKNKHMNADT DYSIAEAAFN KGETAMTING PWAWSNIDTS KV NYGVTVL PTFKGQPSKP FVGVLSAGIN AASPNKELAK EFLENYLLTD EGLEAVNKDK PLGAVALKSY EEELAKDPRI AAT MENAQK GEIMPNIPQM SAFWYAVRTA VINAASGRQT VDEALKDAQT IIELYRQSLE IISRYLREQA TGAADTAPMG RSGA TSRKA LETLRRVGDG VQRNHETAFQ GMLRKLDIKN EDDVKSLSRV MIHVFSDGVT NWGRIVTLIS FGAFVAKHLK TINQE SAIE PLAESITDVL VRTKRDWLVK QRGWDGFVEF F |
-Macromolecule #2: Bcl-2 homologous antagonist/killer
| Macromolecule | Name: Bcl-2 homologous antagonist/killer / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.366679 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: STMGQVGRQL AIIGDDINRR Y UniProtKB: Bcl-2 homologous antagonist/killer |
-Macromolecule #3: Synthetic antibody, Fab fragment, Heavy Chain
| Macromolecule | Name: Synthetic antibody, Fab fragment, Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.534371 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFNLS SSSIHWVRQA PGKGLEWVAS IYSYYGSTSY ADSVKGRFTI SADTSKNTAY LQMNSLRAE DTAVYYCARE YHSYWSYSWW PRVGLDYWGQ GTLVTVSSAS TKGPSVFPLA PASKSAAAAT AALGCLVKDY F PEPVTVSW ...String: EVQLVESGGG LVQPGGSLRL SCAASGFNLS SSSIHWVRQA PGKGLEWVAS IYSYYGSTSY ADSVKGRFTI SADTSKNTAY LQMNSLRAE DTAVYYCARE YHSYWSYSWW PRVGLDYWGQ GTLVTVSSAS TKGPSVFPLA PASKSAAAAT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SC |
-Macromolecule #4: Synthetic antibody, Fab fragment, Light Chain
| Macromolecule | Name: Synthetic antibody, Fab fragment, Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.109719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AQMTQSPSSL SASVGDRVTI TCRASQSVSS AVAWYQQKPG KAPKLLIYSA SSLYSGVPSR FSGSRSGTDF TLTISSLQPE DFATYYCQQ ASLTALLTFG QGTKVEIKRT VAAPSVFIFP PSDSQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: AQMTQSPSSL SASVGDRVTI TCRASQSVSS AVAWYQQKPG KAPKLLIYSA SSLYSGVPSR FSGSRSGTDF TLTISSLQPE DFATYYCQQ ASLTALLTFG QGTKVEIKRT VAAPSVFIFP PSDSQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)