[English] 日本語
 Yorodumi
Yorodumi- EMDB-41480: nhTMEM16 R432A mutant in lipid nanodiscs with MSP2N2 scaffold pro... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
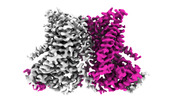
| Title | nhTMEM16 R432A mutant in lipid nanodiscs with MSP2N2 scaffold protein in the presence of Ca2+ | |||||||||
 Map data Map data | Primary map used for model buidling | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / lipid scramblase / TMEM16 / LIPID TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationcortical endoplasmic reticulum / chloride channel activity / metal ion binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Fusarium vanettenii 77-13-4 (fungus) Fusarium vanettenii 77-13-4 (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.64 Å | |||||||||
 Authors Authors | Feng Z / Accardi A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of closed groove scrambling by a TMEM16 protein. Authors: Zhang Feng / Omar E Alvarenga / Alessio Accardi /  Abstract: Activation of Ca-dependent TMEM16 scramblases induces phosphatidylserine externalization, a key step in multiple signaling processes. Current models suggest that the TMEM16s scramble lipids by ...Activation of Ca-dependent TMEM16 scramblases induces phosphatidylserine externalization, a key step in multiple signaling processes. Current models suggest that the TMEM16s scramble lipids by deforming the membrane near a hydrophilic groove and that Ca dependence arises from the different association of lipids with an open or closed groove. However, the molecular rearrangements underlying groove opening and how lipids reorganize outside the closed groove remain unknown. Here we directly visualize how lipids associate at the closed groove of Ca-bound fungal nhTMEM16 in nanodiscs using cryo-EM. Functional experiments pinpoint lipid-protein interaction sites critical for closed groove scrambling. Structural and functional analyses suggest groove opening entails the sequential appearance of two π-helical turns in the groove-lining TM6 helix and identify critical rearrangements. Finally, we show that the choice of scaffold protein and lipids affects the conformations of nhTMEM16 and their distribution, highlighting a key role of these factors in cryo-EM structure determination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41480.map.gz emd_41480.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41480-v30.xml emd-41480-v30.xml emd-41480.xml emd-41480.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41480_fsc.xml emd_41480_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_41480.png emd_41480.png | 85.8 KB | ||
| Filedesc metadata |  emd-41480.cif.gz emd-41480.cif.gz | 6.1 KB | ||
| Others |  emd_41480_half_map_1.map.gz emd_41480_half_map_1.map.gz emd_41480_half_map_2.map.gz emd_41480_half_map_2.map.gz | 79.3 MB 79.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41480 http://ftp.pdbj.org/pub/emdb/structures/EMD-41480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41480 | HTTPS FTP |
-Related structure data
| Related structure data |  8tppMC  8toiC  8tokC  8tolC  8tpmC  8tpnC  8tpoC  8tpqC  8tprC  8tpsC  8tptC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41480.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41480.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map used for model buidling | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: halfmap2
| File | emd_41480_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap1
| File | emd_41480_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimeric lipid scramblase nhTMEM16
| Entire | Name: Dimeric lipid scramblase nhTMEM16 |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric lipid scramblase nhTMEM16
| Supramolecule | Name: Dimeric lipid scramblase nhTMEM16 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Fusarium vanettenii 77-13-4 (fungus) Fusarium vanettenii 77-13-4 (fungus) |
-Macromolecule #1: Lipid scramblase nhTMEM16
| Macromolecule | Name: Lipid scramblase nhTMEM16 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Fusarium vanettenii 77-13-4 (fungus) Fusarium vanettenii 77-13-4 (fungus) |
| Molecular weight | Theoretical: 83.207945 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPSNLKDFSQ PGSGQESNFG VDFVIHYKVP AAERDEAEAG FVQLIRALTT VGLATEVRHG ENESLLVFVK VASPDLFAKQ VYRARLGDW LHGVRVSAPH NDIAQALQDE PVVEAERLRL IYLMITKPHN EGGAGVTPTN AKWKHVESIF PLHSHSFNKE W IKKWSSKY ...String: GPSNLKDFSQ PGSGQESNFG VDFVIHYKVP AAERDEAEAG FVQLIRALTT VGLATEVRHG ENESLLVFVK VASPDLFAKQ VYRARLGDW LHGVRVSAPH NDIAQALQDE PVVEAERLRL IYLMITKPHN EGGAGVTPTN AKWKHVESIF PLHSHSFNKE W IKKWSSKY TLEQTDIDNI RDKFGESVAF YFAFLRSYFR FLVIPSAFGF GAWLLLGQFS YLYALLCGLW SVVFFEYWKK QE VDLAVQW GVRGVSSIQQ SRPEFEWEHE AEDPITGEPV KVYPPMKRVK TQLLQIPFAL ACVVALGALI VTCNSLEVFI NEV YSGPGK QYLGFLPTIF LVIGTPTISG VLMGAAEKLN AMENYATVDA HDAALIQKQF VLNFMTSYMA LFFTAFVYIP FGHI LHPFL NFWRATAQTL TFSEKELPTR EFQINPAAIS NQMFYFTVTA QIVNFATEVV VPYIKQQAFQ KAKQLKSGSK VQEDH EEEA EFLQRVREEC TLEEYDVSGD YREMVMQFGY VAMFSVAWPL AACCFLVNNW VELRSDALKI AISSRRPIPW RTDSIG PWL TALSFLSWLG SITSSAIVYL CSNSKNGTQG EASPLKAWGL LLSILFAEHF YLVVQLAVRF VLSKLDSPGL QKERKER FQ TKKRLLQENL GQDAAEEAAA PGIEHSEKIT REALEEEARQ ASIRGHGTPE EMFWQRQRGM QETIEIGRRM IEQQLAAG K NGKKSAPAVP SEKASA UniProtKB: Plasma membrane channel protein |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 9493 / Average exposure time: 3.4 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)