[English] 日本語
 Yorodumi
Yorodumi- EMDB-4117: cryoEM structure of crenactin double helical filament at 3.8A res... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4117 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of crenactin double helical filament at 3.8A resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / cytoskeleton / hydrolase activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Pyrobaculum calidifontis (archaea) / Pyrobaculum calidifontis (archaea) /   Pyrobaculum calidifontis (strain JCM 11548 / VA1) (archaea) Pyrobaculum calidifontis (strain JCM 11548 / VA1) (archaea) | |||||||||
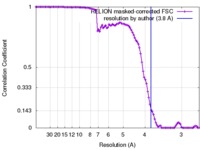
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Izore T / Lowe J | |||||||||
 Citation Citation |  Journal: Elife / Year: 2016 Journal: Elife / Year: 2016Title: Crenactin forms actin-like double helical filaments regulated by arcadin-2. Authors: Thierry Izoré / Danguole Kureisaite-Ciziene / Stephen H McLaughlin / Jan Löwe /  Abstract: The similarity of eukaryotic actin to crenactin, a filament-forming protein from the crenarchaeon supports the theory of a common origin of Crenarchaea and Eukaryotes. Monomeric structures of ...The similarity of eukaryotic actin to crenactin, a filament-forming protein from the crenarchaeon supports the theory of a common origin of Crenarchaea and Eukaryotes. Monomeric structures of crenactin and actin are similar, although their filament architectures were suggested to be different. Here we report that crenactin forms double helical filaments that show exceptional similarity to eukaryotic F-actin. With cryo-electron microscopy and helical reconstruction we solved the structure of the crenactin filament to 3.8 Å resolution. When forming double filaments, the 'hydrophobic plug' loop in crenactin rearranges. Arcadin-2, also encoded by the arcade gene cluster, binds tightly with its C-terminus to the hydrophobic groove of crenactin. Binding is reminiscent of eukaryotic actin modulators such as cofilin and thymosin β4 and arcadin-2 is a depolymeriser of crenactin filaments. Our work further supports the theory of shared ancestry of Eukaryotes and Crenarchaea. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4117.map.gz emd_4117.map.gz | 78.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4117-v30.xml emd-4117-v30.xml emd-4117.xml emd-4117.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4117_fsc.xml emd_4117_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_4117.png emd_4117.png | 38.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4117 http://ftp.pdbj.org/pub/emdb/structures/EMD-4117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4117 | HTTPS FTP |
-Validation report
| Summary document |  emd_4117_validation.pdf.gz emd_4117_validation.pdf.gz | 309.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4117_full_validation.pdf.gz emd_4117_full_validation.pdf.gz | 308.9 KB | Display | |
| Data in XML |  emd_4117_validation.xml.gz emd_4117_validation.xml.gz | 11 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4117 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4117 | HTTPS FTP |
-Related structure data
| Related structure data |  5mw1MC  5ly3C  5ly5C  5ly4 M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4117.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4117.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : crenactin
| Entire | Name: crenactin |
|---|---|
| Components |
|
-Supramolecule #1: crenactin
| Supramolecule | Name: crenactin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Pyrobaculum calidifontis (archaea) Pyrobaculum calidifontis (archaea) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Actin/actin family protein
| Macromolecule | Name: Actin/actin family protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrobaculum calidifontis (strain JCM 11548 / VA1) (archaea) Pyrobaculum calidifontis (strain JCM 11548 / VA1) (archaea)Strain: JCM 11548 / VA1 |
| Molecular weight | Theoretical: 48.425484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGVISDAYRL KYTFGVDFGT SYVKYGPITL NEPKMVQTRG LFLRDLPESV KMRIPPDVLA RGLVVGDEEV RKYLSSVRDV QRNLKYPLK DGVARRDDEE AWRVLKELAR YTLAQFPVSD PEFAGWLVAV ALSALAPDYM YKAIFDIYDE LASEFKIYAV T ILPQPLAV ...String: MGVISDAYRL KYTFGVDFGT SYVKYGPITL NEPKMVQTRG LFLRDLPESV KMRIPPDVLA RGLVVGDEEV RKYLSSVRDV QRNLKYPLK DGVARRDDEE AWRVLKELAR YTLAQFPVSD PEFAGWLVAV ALSALAPDYM YKAIFDIYDE LASEFKIYAV T ILPQPLAV AIAENAVNCV IVEGGHGNIQ VAPISFALIR EGLVALNRGG AEANAITREI LKDIGYSDIA REEYAVEVVK RA VGLVPRR LKEAIRAAKS DPDRFVTKVR LSPVVEVEIP REYAWTRFLI GEIVFDPNHE EIKSYIEQSR LRIENAVIGD VTL YGEMDV ASAIITSLRN VSVEIQERVA SQIILSGGAF SWRVPPGMED VAADSVTRVK IALEEKSPAL ASKVEVRLVS EPQY SVWRG AVIYGYALPL SLEWSDTTRE GWRFPRR |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number real images: 1474 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Target criteria: R-factor & stereochemistry |
|---|---|
| Output model |  PDB-5mw1: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)