[English] 日本語
 Yorodumi
Yorodumi- EMDB-40716: CryoEM structure of a therapeutic antibody (favezelimab) bound to... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lymphocyte activation gene 3 protein / favezelimab-LAG3 complex / cryoEM / Single particle reconstruction. / IMMUNOSUPPRESSANT | |||||||||
| Function / homology |  Function and homology information Function and homology informationplasmacytoid dendritic cell activation / negative regulation of regulatory T cell differentiation / positive regulation of natural killer cell mediated cytotoxicity / MHC class II protein binding / negative regulation of interleukin-2 production / natural killer cell mediated cytotoxicity / regulation of immune response / antigen binding / MHC class II antigen presentation / T cell activation ...plasmacytoid dendritic cell activation / negative regulation of regulatory T cell differentiation / positive regulation of natural killer cell mediated cytotoxicity / MHC class II protein binding / negative regulation of interleukin-2 production / natural killer cell mediated cytotoxicity / regulation of immune response / antigen binding / MHC class II antigen presentation / T cell activation / transmembrane signaling receptor activity / adaptive immune response / cell surface receptor signaling pathway / innate immune response / external side of plasma membrane / cell surface / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.53 Å | |||||||||
 Authors Authors | Mishra AK / Shahid S / Karade SS / Mariuzza RA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker. Authors: Arjun K Mishra / Salman Shahid / Sharanbasappa S Karade / Pragati Agnihotri / Alexander Kolesnikov / S Saif Hasan / Roy A Mariuzza /  Abstract: Lymphocyte activation gene 3 protein (LAG3) is an inhibitory receptor that is upregulated on exhausted T cells in tumors. LAG3 is a major target for cancer immunotherapy with many anti-LAG3 ...Lymphocyte activation gene 3 protein (LAG3) is an inhibitory receptor that is upregulated on exhausted T cells in tumors. LAG3 is a major target for cancer immunotherapy with many anti-LAG3 antibodies in clinical trials. However, there is no structural information on the epitopes recognized by these antibodies. We determined the single-particle cryoEM structure of a therapeutic antibody (favezelimab) bound to LAG3 to 3.5 Å resolution, revealing that favezelimab targets the LAG3-binding site for MHC class II, its canonical ligand. The small size of the complex between the conventional (monovalent) Fab of favezelimab and LAG3 (∼100 kDa) presented a challenge for cryoEM. Accordingly, we engineered a bivalent version of Fab favezelimab that doubled the size of the Fab-LAG3 complex and conferred a highly identifiable shape to the complex that facilitated particle selection and orientation for image processing. This study establishes bivalent Fabs as new fiducial markers for cryoEM analysis of small proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40716.map.gz emd_40716.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40716-v30.xml emd-40716-v30.xml emd-40716.xml emd-40716.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40716.png emd_40716.png | 80.6 KB | ||
| Filedesc metadata |  emd-40716.cif.gz emd-40716.cif.gz | 6.3 KB | ||
| Others |  emd_40716_half_map_1.map.gz emd_40716_half_map_1.map.gz emd_40716_half_map_2.map.gz emd_40716_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40716 http://ftp.pdbj.org/pub/emdb/structures/EMD-40716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40716 | HTTPS FTP |
-Related structure data
| Related structure data |  8sr0MC  6wkmC  8fwhC  8so3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40716.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40716.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||
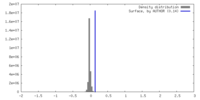
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40716_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
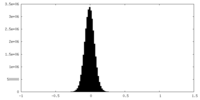
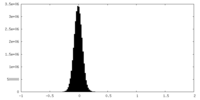
| Density Histograms |
-Half map: #1
| File | emd_40716_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
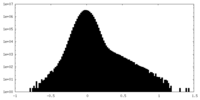
| Density Histograms |
- Sample components
Sample components
-Entire : therapeutic antibody (favezelimab) bound to human LAG3
| Entire | Name: therapeutic antibody (favezelimab) bound to human LAG3 |
|---|---|
| Components |
|
-Supramolecule #1: therapeutic antibody (favezelimab) bound to human LAG3
| Supramolecule | Name: therapeutic antibody (favezelimab) bound to human LAG3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Lymphocyte activation gene 3 protein
| Macromolecule | Name: Lymphocyte activation gene 3 protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.50827 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MWEAQFLGLL FLQPLWVAPV KPLQPGAEVP VVWAQEGAPA QLPCSPTIPL QDLSLLRRAG VTWQHQPDSG PPAAAPGHPL APGPHPAAP SSWGPRPRRY TVLSVGPGGL RSGRLPLQPR VQLDERGRQR GDFSLWLRPA RRADAGEYRA AVHLRDRALS C RLRLRLGQ ...String: MWEAQFLGLL FLQPLWVAPV KPLQPGAEVP VVWAQEGAPA QLPCSPTIPL QDLSLLRRAG VTWQHQPDSG PPAAAPGHPL APGPHPAAP SSWGPRPRRY TVLSVGPGGL RSGRLPLQPR VQLDERGRQR GDFSLWLRPA RRADAGEYRA AVHLRDRALS C RLRLRLGQ ASMTASPPGS LRASDWVILN CSFSRPDRPA SVHWFRNRGQ GRVPVRESPH HHLAESFLFL PQVSPMDSGP WG CILTYRD GFNVSIMYNL TVLGLEPPTP LTVYAGAGSR VGLPCRLPAG VGTRSFLTAK WTPPGGGPDL LVTGDNGDFT LRL EDVSQA QAGTYTCHIH LQEQQLNATV TLAIITVTPK SFGSPGSLGK LLCEVTPVSG QERFVWSSLD TPSQRSFSGP WLEA QEAQL LSQPWQCQLY QGERLLGAAV YFTELSSPGA QRSGRAPGAL PAGHLLLFLI LGVLSLLLLV TGAFGFHLWR RQWRP RRFS ALEQGIHPPQ AQSKIEELEQ EPEPEPEPEP EPEPEPEPEQ L UniProtKB: Lymphocyte activation gene 3 protein |
-Macromolecule #2: favezelimab Fab heavy chain
| Macromolecule | Name: favezelimab Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.245727 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWTWIFLFF LSGTAGVLSE VLLLQSGPEL VKPGTSVKIP CKASGYTFTD YNVDWVKQRH GKGLEWIGDI NPNNGGTIYS QKFKGKATL TVDKSSSTAF MELRSLTSED TAVYFCARNY RWFGAMDHWG QGTSVTVSST KGPSVFPLAP SSKSTSGGTA A LGCLVKDY ...String: MGWTWIFLFF LSGTAGVLSE VLLLQSGPEL VKPGTSVKIP CKASGYTFTD YNVDWVKQRH GKGLEWIGDI NPNNGGTIYS QKFKGKATL TVDKSSSTAF MELRSLTSED TAVYFCARNY RWFGAMDHWG QGTSVTVSST KGPSVFPLAP SSKSTSGGTA A LGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKRVEPKS CD KTAGWSH PQFEK |
-Macromolecule #3: favezelimab Fab light chain
| Macromolecule | Name: favezelimab Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.915627 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METDTILLWV LLLWVPGSTG DIVLTQSPAS LAVSPGQRAT ISCKASQSLD YEGDSDMNWY QQKPGQPPRL LISGASNLES GIPARFSGS GSGTDFTVNI HPVEEEDAAT YYCQQSTEDP RTFGGGTKLE IKRTVAAPSV FIFPPSDEQL KSGTASVVCL L NNFYPREA ...String: METDTILLWV LLLWVPGSTG DIVLTQSPAS LAVSPGQRAT ISCKASQSLD YEGDSDMNWY QQKPGQPPRL LISGASNLES GIPARFSGS GSGTDFTVNI HPVEEEDAAT YYCQQSTEDP RTFGGGTKLE IKRTVAAPSV FIFPPSDEQL KSGTASVVCL L NNFYPREA KYQWKVDNAL QSGNSQESVT EQDSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9508 / Average exposure time: 3.2 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.53 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 171144 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)