+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | structure of SPG11-SPG15 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphagosome-lysosome fusion involved in apoptotic cell clearance / walking behavior / corticospinal tract morphogenesis / regulation of store-operated calcium entry / localization within membrane / autophagosome organization / phosphatidylinositol-3-phosphate binding / vesicle transport along microtubule / axon extension / axo-dendritic transport ...phagosome-lysosome fusion involved in apoptotic cell clearance / walking behavior / corticospinal tract morphogenesis / regulation of store-operated calcium entry / localization within membrane / autophagosome organization / phosphatidylinositol-3-phosphate binding / vesicle transport along microtubule / axon extension / axo-dendritic transport / cholesterol efflux / motor neuron apoptotic process / lysosome organization / synaptic vesicle transport / neuromuscular junction development / motor behavior / mitotic cytokinesis / skeletal muscle fiber development / axonogenesis / regulation of cytokinesis / protein catabolic process / double-strand break repair via homologous recombination / memory / protein import into nucleus / late endosome / cytoplasmic vesicle / midbody / chemical synaptic transmission / early endosome / lysosome / axon / lysosomal membrane / dendrite / synapse / centrosome / protein kinase binding / nucleolus / endoplasmic reticulum / zinc ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.02 Å | |||||||||
 Authors Authors | Su M-Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural basis for membrane remodeling by the AP5-SPG11-SPG15 complex. Authors: Xinyi Mai / Yang Wang / Xi Wang / Ming Liu / Fei Teng / Zheng Liu / Ming-Yuan Su / Goran Stjepanovic /  Abstract: The human spastizin (spastic paraplegia 15, SPG15) and spatacsin (spastic paraplegia 11, SPG11) complex is involved in the formation of lysosomes, and mutations in these two proteins are linked with ...The human spastizin (spastic paraplegia 15, SPG15) and spatacsin (spastic paraplegia 11, SPG11) complex is involved in the formation of lysosomes, and mutations in these two proteins are linked with hereditary autosomal-recessive spastic paraplegia. SPG11-SPG15 can cooperate with the fifth adaptor protein complex (AP5) involved in membrane sorting of late endosomes. We employed cryogenic-electron microscopy and in silico predictions to investigate the structural assemblies of the SPG11-SPG15 and AP5-SPG11-SPG15 complexes. The W-shaped SPG11-SPG15 intertwined in a head-to-head fashion, and the N-terminal region of SPG11 is required for AP5 complex interaction and assembly. The AP5 complex is in a super-open conformation. Our findings reveal that the AP5-SPG11-SPG15 complex can bind PI3P molecules, sense membrane curvature and drive membrane remodeling in vitro. These studies provide insights into the structure and function of the spastic paraplegia AP5-SPG11-SPG15 complex, which is essential for the initiation of autolysosome tubulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39096.map.gz emd_39096.map.gz | 967.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39096-v30.xml emd-39096-v30.xml emd-39096.xml emd-39096.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39096.png emd_39096.png | 74.4 KB | ||
| Filedesc metadata |  emd-39096.cif.gz emd-39096.cif.gz | 9.1 KB | ||
| Others |  emd_39096_additional_1.map.gz emd_39096_additional_1.map.gz emd_39096_half_map_1.map.gz emd_39096_half_map_1.map.gz emd_39096_half_map_2.map.gz emd_39096_half_map_2.map.gz | 1.7 GB 1.8 GB 1.8 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39096 http://ftp.pdbj.org/pub/emdb/structures/EMD-39096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39096 | HTTPS FTP |
-Related structure data
| Related structure data |  8yadMC  8yabC  8yahC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39096.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39096.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: deepemhancer map
| File | emd_39096_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepemhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
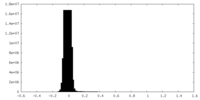
| Density Histograms |
-Half map: #2
| File | emd_39096_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
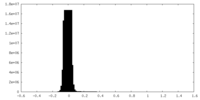
| Density Histograms |
-Half map: #1
| File | emd_39096_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SPG11-SPG15 complex
| Entire | Name: SPG11-SPG15 complex |
|---|---|
| Components |
|
-Supramolecule #1: SPG11-SPG15 complex
| Supramolecule | Name: SPG11-SPG15 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 568 KDa |
-Macromolecule #1: Spatacsin
| Macromolecule | Name: Spatacsin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 279.182594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAEEGVASA ASAGGSWGTA AMGRVLPMLL VPVPAEAMGQ LGSRAQLRTQ PEALGSLTAA GSLQVLSLTP GSRGGGRCCL EGPFWHFLW EDSRNSSTPT EKPKLLALGE NYELLIYEFN LKDGRCDATI LYSCSREALQ KLIDDQDISI SLLSLRILSF H NNTSLLFI ...String: MAAEEGVASA ASAGGSWGTA AMGRVLPMLL VPVPAEAMGQ LGSRAQLRTQ PEALGSLTAA GSLQVLSLTP GSRGGGRCCL EGPFWHFLW EDSRNSSTPT EKPKLLALGE NYELLIYEFN LKDGRCDATI LYSCSREALQ KLIDDQDISI SLLSLRILSF H NNTSLLFI NKCVILHIIF PERDAAIRVL NCFTLPLPAQ AVDMIIDTQL CRGILFVLSS LGWIYIFDVV DGTYVAHVDL AL HKEDMCN EQQQEPAKIS SFTSLKVSQD LDVAVIVSSS NSAVALNLNL YFRQHPGHLL CERILEDLPI QGPKGVDEDD PVN SAYNMK LAKFSFQIDR SWKAQLSSLN ETIKNSKLEV SCCAPWFQDI LHLESPESGN HSTSVQSWAF IPQDIMHGQY NVLQ KDHAK TSDPGRSWKI MHISEQEEPI ELKCVSVTGF TALFTWEVER MGYTITLWDL ETQGMQCFSL GTKCIPVDSS GDQQL CFVL TENGLSLILF GLTQEEFLNR LMIHGSASTV DTLCHLNGWG RCSIPIHALE AGIENRQLDT VNFFLKSKEN LFNPSS KSS VSDQFDHLSS HLYLRNVEEL IPALDLLCSA IRESYSEPQS KHFSEQLLNL TLSFLNNQIK ELFIHTEELD EHLQKGV NI LTSYINELRT FMIKFPWKLT DAIDEYDVHE NVPKVKESNI WKKLSFEEVI ASAILNNKIP EAQTFFRIDS HSAQKLEE L IGIGLNLVFD NLKKNNIKEA SELLKNMGFD VKGQLLKICF YTTNKNIRDF LVEILKEKNY FSEKEKRTID FVHQVEKLY LGHFQENMQI QSFPRYWIKE QDFFKHKSVL DSFLKYDCKD EFNKQDHRIV LNWALWWDQL TQESILLPRI SPEEYKSYSP EALWRYLTA RHDWLNIILW IGEFQTQHSY ASLQQNKWPL LTVDVINQNT SCNNYMRNEI LDKLARNGVF LASELEDFEC F LLRLSRIG GVIQDTLPVQ NYKTKEGWDF HSQFILYCLE HSLQHLLYVY LDCYKLSPEN CPFLEKKELH EAHPWFEFLV QC RQVASNL TDPKLIFQAS LANAQILIPT NQASVSSMLL EGHTLLALAT TMYSPGGVSQ VVQNEENENC LKKVDPQLLK MAL TPYPKL KTALFPQCTP PSVLPSDITI YHLIQSLSPF DPSRLFGWQS ANTLAIGDAW SHLPHFSSPD LVNKYAIVER LNFA YYLHN GRPSFAFGTF LVQELIKSKT PKQLIQQVGN EAYVIGLSSF HIPSIGAACV CFLELLGLDS LKLRVDMKVA NIILS YKCR NEDAQYSFIR ESVAEKLSKL ADGEKTTTEE LLVLLEEGTW NSIQQQEIKR LSSESSSQWA LVVQFCRLHN MKLSIS YLR ECAKANDWLQ FIIHSQLHNY HPAEVKSLIQ YFSPVIQDHL RLAFENLPSV PTSKMDSDQV CNKCPQELQG SKQEMTD LF EILLQCSEEP DSWHWLLVEA VKQQAPILSV LASCLQGASA ISCLCVWIIT SVEDNVATEA MGHIQDSTED HTWNLEDL S VIWRTLLTRQ KSKTLIRGFQ LFFKDSPLLL VMEMYELCMF FRNYKEAEAK LLEFQKSLET LNTAATKVHP VIPAMWLED QVCFLLKLML QQCKTQYELG KLLQLFVERE HLFSDGPDVK KLCILCQILK DTSIAINHTI ITSYSIENLQ HECRSILERL QTDGQFALA RRVAELAELP VDNLVIKEIT QEMQTLKHIE QWSLKQARID FWKKCHENFK KNSISSKAAS SFFSTQAHVA C EHPTGWSS MEERHLLLTL AGHWLAQEDV VPLDKLEELE KQIWLCRITQ HTLGRNQEET EPRFSRQIST SGELSFDSLA SE FSFSKLA ALNTSKYLEL NSLPSKETCE NRLDWKEQES LNFLIGRLLD DGCVHEASRV CRYFHFYNPD VALVLHCRAL ASG EASMED LHPEIHALLQ SAELLEEEAP DIPLRRVHST SSLDSQKFVT VPSSNEVVTN LEVLTSKCLH GKNYCRQVLC LYDL AKELG CSYTDVAAQD GEAMLRKILA SQQPDRCKRA QAFISTQGLK PDTVAELVAE EVTRELLTSS QGTGHKQMFN PTEES QTFL QLTTLCQDRT LVGMKLLDKI SSVPHGELSC TTELLILAHH CFTLTCHMEG IIRVLQAAHM LTDNHLAPSE EYGLVV RLL TGIGRYNEMT YIFDLLHKKH YFEVLMRKKL DPSGTLKTAL LDYIKRCRPG DSEKHNMIAL CFSMCREIGE NHEAAAR IQ LKLIESQPWE DSLKDGHQLK QLLLKALTLM LDAAESYAKD SCVRQAQHCQ RLTKLITLQI HFLNTGQNTM LINLGRHK L MDCILALPRF YQASIVAEAY DFVPDWAEIL YQQVILKGDF NYLEEFKQQR LLKSSIFEEI SKKYKQHQPT DMVMENLKK LLTYCEDVYL YYKLAYEHKF YEIVNVLLKD PQTGCCLKDM LAG UniProtKB: Spatacsin |
-Macromolecule #2: Zinc finger FYVE domain-containing protein 26
| Macromolecule | Name: Zinc finger FYVE domain-containing protein 26 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 284.943906 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNHPFGKEEA ASQKQLFGFF CECLRRGEWE LAQACVPQLQ EGQGDIPKRV EDILQALVVC PNLLRCGQDI NPQRVAWVWL LVLEKWLAR EKKLLPVVFR RKLEFLLLSE DLQGDIPENI LEELYETLTQ GAVGHVPDGN PRRESWTPRL SSEAVSVLWD L LRQSPQPA ...String: MNHPFGKEEA ASQKQLFGFF CECLRRGEWE LAQACVPQLQ EGQGDIPKRV EDILQALVVC PNLLRCGQDI NPQRVAWVWL LVLEKWLAR EKKLLPVVFR RKLEFLLLSE DLQGDIPENI LEELYETLTQ GAVGHVPDGN PRRESWTPRL SSEAVSVLWD L LRQSPQPA QALLELLLEE DDGTGLCHWP LQNALVDLIR KALRALQGPD SVPPGVVDAI YGALRTLRCP AEPLGVELHL LC EELLEAC RTEGSPLREE RLLSCLLHKA SRGLLSLYGH TYAEKVTEKP PRATASGKVS PDHLDPERAM LALFSNPNPA EAW KVAYFY CLSNNKHFLE QILVTALTLL KEEDFPNLGC LLDREFRPLS CLLVLLGWTH CQSLESAKRL LQTLHRTQGP GCDE LLRDA CDGLWAHLEV LEWCIQQSSN PIPKRDLLYH LHGGDSHSVL YTLHHLTNLP ALREEDVLKL LQKVPAKDPQ QEPDA VDAP VPEHLSQCQN LTLYQGFCAM KYAIYALCVN SHQHSQCQDC KDSLSEDLAS ATEPANDSLS SPGAANLFST YLARCQ QYL CSIPDSLCLE LLENIFSLLL ITSADLHPEP HLPEDYAEDD DIEGKSPSGL RSPSESPQHI AHPERKSERG SLGVPKT LA YTMPSHVKAE PKDSYPGPHR HSFLDLKHFT SGISGFLADE FAIGAFLRLL QEQLDEISSR SPPEKPKQES QSCSGSRD G LQSRLHRLSK VVSEAQWRHK VVTSNHRSEE QPSRRYQPAT RHPSLRRGRR TRRSQADGRD RGSNPSLEST SSELSTSTS EGSLSAMSGR NELHSRLHPH PQSSLIPMMF SPPESLLASC ILRGNFAEAH QVLFTFNLKS SPSSGELMFM ERYQEVIQEL AQVEHKIEN QNSDAGSSTI RRTGSGRSTL QAIGSAAAAG MVFYSISDVT DKLLNTSGDP IPMLQEDFWI STALVEPTAP L REVLEDLS PPAMAAFDLA CSQCQLWKTC KQLLETAERR LNSSLERRGR RIDHVLLNAD GIRGFPVVLQ QISKSLNYLL MS ASQTKSE SVEEKGGGPP RCSITELLQM CWPSLSEDCV ASHTTLSQQL DQVLQSLREA LELPEPRTPP LSSLVEQAAQ KAP EAEAHP VQIQTQLLQK NLGKQTPSGS RQMDYLGTFF SYCSTLAAVL LQSLSSEPDH VEVKVGNPFV LLQQSSSQLV SHLL FERQV PPERLAALLA QENLSLSVPQ VIVSCCCEPL ALCSSRQSQQ TSSLLTRLGT LAQLHASHCL DDLPLSTPSS PRTTE NPTL ERKPYSSPRD SSLPALTSSA LAFLKSRSKL LATVACLGAS PRLKVSKPSL SWKELRGRRE VPLAAEQVAR ECERLL EQF PLFEAFLLAA WEPLRGSLQQ GQSLAVNLCG WASLSTVLLG LHSPIALDVL SEAFEESLVA RDWSRALQLT EVYGRDV DD LSSIKDAVLS CAVACDKEGW QYLFPVKDAS LRSRLALQFV DRWPLESCLE ILAYCISDTA VQEGLKCELQ RKLAELQV Y QKILGLQSPP VWCDWQTLRS CCVEDPSTVM NMILEAQEYE LCEEWGCLYP IPREHLISLH QKHLLHLLER RDHDKALQL LRRIPDPTMC LEVTEQSLDQ HTSLATSHFL ANYLTTHFYG QLTAVRHREI QALYVGSKIL LTLPEQHRAS YSHLSSNPLF MLEQLLMNM KVDWATVAVQ TLQQLLVGQE IGFTMDEVDS LLSRYAEKAL DFPYPQREKR SDSVIHLQEI VHQAADPETL P RSPSAEFS PAAPPGISSI HSPSLRERSF PPTQPSQEFV PPATPPARHQ WVPDETESIC MVCCREHFTM FNRRHHCRRC GR LVCSSCS TKKMVVEGCR ENPARVCDQC YSYCNKDVPE EPSEKPEALD SSKNESPPYS FVVRVPKADE VEWILDLKEE ENE LVRSEF YYEQAPSASL CIAILNLHRD SIACGHQLIE HCCRLSKGLT NPEVDAGLLT DIMKQLLFSA KMMFVKAGQS QDLA LCDSY ISKVDVLNIL VAAAYRHVPS LDQILQPAAV TRLRNQLLEA EYYQLGVEVS TKTGLDTTGA WHAWGMACLK AGNLT AARE KFSRCLKPPF DLNQLNHGSR LVQDVVEYLE STVRPFVSLQ DDDYFATLRE LEATLRTQSL SLAVIPEGKI MNNTYY QEC LFYLHNYSTN LAIISFYVRH SCLREALLHL LNKESPPEVF IEGIFQPSYK SGKLHTLENL LESIDPTLES WGKYLIA AC QHLQKKNYYH ILYELQQFMK DQVRAAMTCI RFFSHKAKSY TELGEKLSWL LKAKDHLKIY LQETSRSSGR KKTTFFRK K MTAADVSRHM NTLQLQMEVT RFLHRCESAG TSQITTLPLP TLFGNNHMKM DVACKVMLGG KNVEDGFGIA FRVLQDFQL DAAMTYCRAA RQLVEKEKYS EIQQLLKCVS ESGMAAKSDG DTILLNCLEA FKRIPPQELE GLIQAIHNDD NKVRAYLICC KLRSAYLIA VKQEHSRATA LVQQVQQAAK SSGDAVVQDI CAQWLLTSHP RGAHGPGSRK UniProtKB: Zinc finger FYVE domain-containing protein 26 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.2386 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)