[English] 日本語
 Yorodumi
Yorodumi- EMDB-32336: Structure of a human glycosylphosphatidylinositol (GPI) transamidase -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32336 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a human glycosylphosphatidylinositol (GPI) transamidase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycosylphosphatidylinositol / transamidase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationGPI-anchor transamidase activity / attachment of GPI anchor to protein / GPI-anchor transamidase complex / GPI anchored protein biosynthesis / GPI anchor biosynthetic process / GPI anchor binding / Attachment of GPI anchor to uPAR / protein retention in ER lumen / Transferases; Transferring nitrogenous groups; Transaminases / regulation of receptor signaling pathway via JAK-STAT ...GPI-anchor transamidase activity / attachment of GPI anchor to protein / GPI-anchor transamidase complex / GPI anchored protein biosynthesis / GPI anchor biosynthetic process / GPI anchor binding / Attachment of GPI anchor to uPAR / protein retention in ER lumen / Transferases; Transferring nitrogenous groups; Transaminases / regulation of receptor signaling pathway via JAK-STAT / neuron differentiation / neuron apoptotic process / cytoplasmic vesicle / centrosome / endoplasmic reticulum membrane / endoplasmic reticulum / mitochondrion / proteolysis / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zhang H / Su J | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structure of human glycosylphosphatidylinositol transamidase. Authors: Hongwei Zhang / Jiawei Su / Bin Li / Yiwei Gao / Mengran Liu / Lingli He / Hao Xu / Yanli Dong / Xuejun Cai Zhang / Yan Zhao /  Abstract: Glycosylphosphatidylinositol (GPI) molecules are complex glycophospholipids and serve as membrane anchors for tethering many proteins to the cell surface. Attaching GPI to the protein in the ...Glycosylphosphatidylinositol (GPI) molecules are complex glycophospholipids and serve as membrane anchors for tethering many proteins to the cell surface. Attaching GPI to the protein in the endoplasmic reticulum (ER) is catalyzed by the transmembrane GPI transamidase (GPIT) complex, which is essential for maturation of the GPI-anchored proteins. The GPIT complex is known to be composed of five subunits: PIGK, PIGU, PIGT, PIGS and GPAA1. Here, we determined the structure of the human GPIT complex at a resolution of 3.1 Å using single-particle cryo-EM, elucidating its overall assembly. The PIGK subunit functions as the catalytic component, in which we identified a C206-H164-N58 triad that is critical for the transamination reaction. Transmembrane helices constitute a widely opened cleft, which is located underneath PIGK, serving as a GPI substrate-binding site. The ubiquitin E3 ligase RNF121 is visualized at the back of the complex and probably serves as a quality control factor for the GPIT complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32336.map.gz emd_32336.map.gz | 13.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32336-v30.xml emd-32336-v30.xml emd-32336.xml emd-32336.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32336.png emd_32336.png | 172.3 KB | ||
| Filedesc metadata |  emd-32336.cif.gz emd-32336.cif.gz | 7.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32336 http://ftp.pdbj.org/pub/emdb/structures/EMD-32336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32336 | HTTPS FTP |
-Related structure data
| Related structure data |  7w72MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32336.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32336.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human glycosylphosphatidylinositol transamidase
| Entire | Name: Human glycosylphosphatidylinositol transamidase |
|---|---|
| Components |
|
-Supramolecule #1: Human glycosylphosphatidylinositol transamidase
| Supramolecule | Name: Human glycosylphosphatidylinositol transamidase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphatidylinositol glycan anchor biosynthesis class U protein
| Macromolecule | Name: Phosphatidylinositol glycan anchor biosynthesis class U protein type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.448422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAPLVLVLV VAVTVRAALF RSSLAEFISE RVEVVSPLSS WKRVVEGLSL LDLGVSPYSG AVFHETPLII YLFHFLIDYA ELVFMITDA LTAIALYFAI QDFNKVVFKK QKLLLELDQY APDVAELIRT PMEMRYIPLK VALFYLLNPY TILSCVAKST C AINNTLIA ...String: MAAPLVLVLV VAVTVRAALF RSSLAEFISE RVEVVSPLSS WKRVVEGLSL LDLGVSPYSG AVFHETPLII YLFHFLIDYA ELVFMITDA LTAIALYFAI QDFNKVVFKK QKLLLELDQY APDVAELIRT PMEMRYIPLK VALFYLLNPY TILSCVAKST C AINNTLIA FFILTTIKGS AFLSAIFLAL ATYQSLYPLT LFVPGLLYLL QRQYIPVKMK SKAFWIFSWE YAMMYVGSLV VI ICLSFFL LSSWDFIPAV YGFILSVPDL TPNIGLFWYF FAEMFEHFSL FFVCVFQINV FFYTIPLAIK LKEHPIFFMF IQI AVIAIF KSYPTVGDVA LYMAFFPVWN HLYRFLRNIF VLTCIIIVCS LLFPVLWHLW IYAGSANSNF FYAITLTFNV GQIL LISDY FYAFLRREYY LTHGL UniProtKB: GPI-anchor transamidase component PIGU |
-Macromolecule #2: GPI transamidase component PIG-S
| Macromolecule | Name: GPI transamidase component PIG-S / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.09782 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AATHLEVARG KRAALFFAAV AIVLGLPLWW KTTETYRASL PYSQISGLNA LQLRLMVPVT VVFTRESVPL DDQAKLPFTV VHEREIPLK YKMKIKCRFQ KAYRRALDHE EEALSSGSVQ EAEAMLDEPQ EQAEGSLTVY VISEHSSLLP QDMASYIGPK R TAVVRGIM ...String: AATHLEVARG KRAALFFAAV AIVLGLPLWW KTTETYRASL PYSQISGLNA LQLRLMVPVT VVFTRESVPL DDQAKLPFTV VHEREIPLK YKMKIKCRFQ KAYRRALDHE EEALSSGSVQ EAEAMLDEPQ EQAEGSLTVY VISEHSSLLP QDMASYIGPK R TAVVRGIM HREAFNIIGR RIVQVAQAMS LTEDVLAAAL ADHLPEDKWS AEKRRPLKSS LGYEITFSLL NPDPKSHDVY WD IEGAVRR YVQPFLNALG AAGNFSVDSQ ILYYAMLGVN PRFDSASSSY YLDMHSLPHV INPVESRLGS SAASLYPVLN FLL YVPELA HSPLYIQDKD GAPVATNAFH SPRWGGIMVY NVDSKTYNAS VLPVRVEVDM VRVMEVFLAQ LRLLFGIAQP QLPP KCLLS GPTSEGLMTW ELDRLLWARS VENLATATTT LTSLAQLLGK ISNIVIKDDV ASEVYKAVAA VQKSAEELAS AHLAS AFVA SQEAVTSSEL AFFDASLLHL LYFPDDQKFA IYIPLFLPMA VPILLSLVK UniProtKB: GPI-anchor transamidase component PIGS |
-Macromolecule #3: GPI transamidase component PIG-T
| Macromolecule | Name: GPI transamidase component PIG-T / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.397691 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ARDSLREELV ITPLPSGDVA ATFQFRTRWD SELQREGVSH YRLFPKALGQ LISKYSLREL HLSFTQGFWR TRYWGPPFLQ APSGAELWV WFQDTVTDVD KSWKELSNVL SGIFCASLNF IDSTNTVTPT ASFKPLGLAN DTDHYFLRYA VLPREVVCTE N LTPWKKLL ...String: ARDSLREELV ITPLPSGDVA ATFQFRTRWD SELQREGVSH YRLFPKALGQ LISKYSLREL HLSFTQGFWR TRYWGPPFLQ APSGAELWV WFQDTVTDVD KSWKELSNVL SGIFCASLNF IDSTNTVTPT ASFKPLGLAN DTDHYFLRYA VLPREVVCTE N LTPWKKLL PCSSKAGLSV LLKADRLFHT SYHSQAVHIR PVCRNARCTS ISWELRQTLS VVFDAFITGQ GKKDWSLFRM FS RTLTEPC PLASESRVYV DITTYNQDNE TLEVHPPPTT TYQDVILGTR KTYAIYDLLD TAMINNSRNL NIQLKWKRPP ENE APPVPF LHAQRYVSGY GLQKGELSTL LYNTHPYRAF PVLLLDTVPW YLRLYVHTLT ITSKGKENKP SYIHYQPAQD RLQP HLLEM LIQLPANSVT KVSIQFERAL LKWTEYTPDP NHGFYVSPSV LSALVPSMVA AKPVDWEESP LFNSLFPVSD GSNYF VRLY TEPLLVNLPT PDFSMPYNVI CLTCTVVAVC YGSFYNLLTR TFH UniProtKB: GPI-anchor transamidase component PIGT |
-Macromolecule #4: GPI-anchor transamidase
| Macromolecule | Name: GPI-anchor transamidase / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.302629 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAVTDSLSRA ATVLATVLLL SFGSVAASHI EDQAEQFFRS GHTNNWAVLV CTSRFWFNYR HVANTLSVYR SVKRLGIPDS HIVLMLADD MACNPRNPKP ATVFSHKNME LNVYGDDVEV DYRSYEVTVE NFLRVLTGRI PPSTPRSKRL LSDDRSNILI Y MTGHGGNG ...String: MAVTDSLSRA ATVLATVLLL SFGSVAASHI EDQAEQFFRS GHTNNWAVLV CTSRFWFNYR HVANTLSVYR SVKRLGIPDS HIVLMLADD MACNPRNPKP ATVFSHKNME LNVYGDDVEV DYRSYEVTVE NFLRVLTGRI PPSTPRSKRL LSDDRSNILI Y MTGHGGNG FLKFQDSEEI TNIELADAFE QMWQKRRYNE LLFIIDTCQG ASMYERFYSP NIMALASSQV GEDSLSHQPD PA IGVHLMD RYTFYVLEFL EEINPASQTN MNDLFQVCPK SLCVSTPGHR TDLFQRDPKN VLITDFFGSV RKVEITTETI KLQ QDSEIM ESSYKEDQMD EKLMEPLKYA EQLPVAQIIH QKPKLKDWHP PGGFILGLWA LIIMVFFKTY GIKHMKFIF UniProtKB: GPI-anchor transamidase |
-Macromolecule #5: Glycosylphosphatidylinositol anchor attachment 1 protein
| Macromolecule | Name: Glycosylphosphatidylinositol anchor attachment 1 protein type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.683836 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGLLSDPVRR RALARLVLRL NAPLCVLSYV AGIAWFLALV FPPLTQRTYM SENAMGSTMV EEQFAGGDRA RAFARDFAAH RKKSGALPV AWLERTMRSV GLEVYTQSFS RKLPFPDETH ERYMVSGTNV YGILRAPRAA STESLVLTVP CGSDSTNSQA V GLLLALAA ...String: MGLLSDPVRR RALARLVLRL NAPLCVLSYV AGIAWFLALV FPPLTQRTYM SENAMGSTMV EEQFAGGDRA RAFARDFAAH RKKSGALPV AWLERTMRSV GLEVYTQSFS RKLPFPDETH ERYMVSGTNV YGILRAPRAA STESLVLTVP CGSDSTNSQA V GLLLALAA HFRGQIYWAK DIVFLVTEHD LLGTEAWLEA YHDVNVTGMQ SSPLQGRAGA IQAAVALELS SDVVTSLDVA VE GLNGQLP NLDLLNLFQT FCQKGGLLCT LQGKLQPEDW TSLDGPLQGL QTLLLMVLRQ ASGRPHGSHG LFLRYRVEAL TLR GINSFR QYKYDLVAVG KALEGMFRKL NHLLERLHQS FFLYLLPGLS RFVSIGLYMP AVGFLLLVLG LKALELWMQL HEAG MGLEE PGGAPGPSVP LPPSQGVGLA SLVAPLLISQ AMGLALYVLP VLGQHVATQH FPVAEAEAVV LTLLAIYAAG LALPH NTHR VVSTQAPDRG WMALKLVALI YLALQLGCIA LTNFSLGFLL ATTMVPTAAL AKPHGPRTLY AALLVLTSPA ATLLGS LFL WRELQEAPLS LAEGWQLFLA ALAQGVLEHH TYGALLFPLL SLGLYPCWLL FWNVLFWK UniProtKB: GPI-anchor transamidase component GPAA1 |
-Macromolecule #7: [2-[[(2~{R})-2-hexanoyloxy-3-[(~{E})-hex-3-enoxy]propoxy]-oxidany...
| Macromolecule | Name: [2-[[(2~{R})-2-hexanoyloxy-3-[(~{E})-hex-3-enoxy]propoxy]-oxidanyl-phosphoryl]oxy-3,4,5,6-tetrakis(oxidanyl)phenyl] (2~{E},4~{E})-hepta-2,4-dienoate type: ligand / ID: 7 / Number of copies: 1 / Formula: 8JY |
|---|---|
| Molecular weight | Theoretical: 622.639 Da |
| Chemical component information | 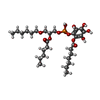 ChemComp-8JY: |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #9: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 9 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 22.0 µm / Nominal defocus min: 12.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















