[English] 日本語
 Yorodumi
Yorodumi- EMDB-32031: Cryo-EM structure of Listeria monocytogenes man-PTS complexed wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32031 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Listeria monocytogenes man-PTS complexed with pediocin PA-1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antimicrobial peptides / bacteriocins / pediocin PA-1 / pediocin-like/class IIa bacteriocins / antibiotic resistance / mannose phosphotransferase / man-PTS / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphoenolpyruvate-dependent sugar phosphotransferase system / killing of cells of another organism / defense response to bacterium / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Listeria monocytogenes (bacteria) / Listeria monocytogenes (bacteria) /  Pediococcus acidilactici (bacteria) Pediococcus acidilactici (bacteria) | |||||||||
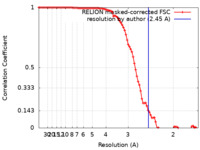
| Method | single particle reconstruction / cryo EM / Resolution: 2.45 Å | |||||||||
 Authors Authors | Wang JW | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Appl Environ Microbiol / Year: 2022 Journal: Appl Environ Microbiol / Year: 2022Title: Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1. Authors: Liyan Zhu / Jianwei Zeng / Chang Wang / Jiawei Wang /  Abstract: Bacteriocins are ribosomally synthesized bacterial antimicrobial peptides that have a narrow spectrum of antibacterial activity against species closely related to the producers. Pediocin-like (or ...Bacteriocins are ribosomally synthesized bacterial antimicrobial peptides that have a narrow spectrum of antibacterial activity against species closely related to the producers. Pediocin-like (or class IIa) bacteriocins (PLBs) exhibit antibacterial activity against several Gram-positive bacterial strains by forming pores in the cytoplasmic membrane of target cells with a specific receptor, the mannose phosphotransferase system (man-PTS). In this study, we report the cryo-electron microscopy structures of man-PTS from Listeria monocytogenes alone and its complex with pediocin PA-1, the first and most extensively studied representative PLB, at resolutions of 3.12 and 2.45 Å, respectively. The structures revealed that the binding of pediocin PA-1 opens the Core domain of man-PTS away from its Vmotif domain, creating a pore through the cytoplasmic membranes of target cells. During this process, the N-terminal β-sheet region of pediocin PA-1 can specifically attach to the extracellular surface of the man-PTS Core domain, whereas the C-terminal half penetrates the membrane and cracks the man-PTS like a wedge. Thus, our findings shed light on a design of novel PLBs that can kill the target pathogenic bacteria. Listeria monocytogenes is a ubiquitous microorganism responsible for listeriosis, a rare but severe disease in humans, who become infected by ingesting contaminated food products (i.e., dairy, meat, fish, and vegetables): the disease has a fatality rate of 33%. Pediocin PA-1 is an important commercial additive used in food production to inhibit species. The mannose phosphotransferase system (man-PTS) is responsible for the sensitivity of Listeria monocytogenes to pediocin PA-1. In this study, we report the cryo-EM structures of man-PTS from Listeria monocytogenes alone and its complex with pediocin PA-1 at resolutions of 3.12 and 2.45 Å, respectively. Our results facilitate the understanding of the mode of action of class IIa bacteriocins as an alternative to antibiotics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32031.map.gz emd_32031.map.gz | 7.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32031-v30.xml emd-32031-v30.xml emd-32031.xml emd-32031.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32031_fsc.xml emd_32031_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32031.png emd_32031.png | 135.8 KB | ||
| Filedesc metadata |  emd-32031.cif.gz emd-32031.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32031 http://ftp.pdbj.org/pub/emdb/structures/EMD-32031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32031 | HTTPS FTP |
-Validation report
| Summary document |  emd_32031_validation.pdf.gz emd_32031_validation.pdf.gz | 424.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32031_full_validation.pdf.gz emd_32031_full_validation.pdf.gz | 424.5 KB | Display | |
| Data in XML |  emd_32031_validation.xml.gz emd_32031_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_32031_validation.cif.gz emd_32031_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32031 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32031 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32031 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32031 | HTTPS FTP |
-Related structure data
| Related structure data |  7vlyMC  7vlxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32031.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32031.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8433 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Listeria monocytogenes man-PTS complexed with pediocin PA-1
| Entire | Name: Listeria monocytogenes man-PTS complexed with pediocin PA-1 |
|---|---|
| Components |
|
-Supramolecule #1: Listeria monocytogenes man-PTS complexed with pediocin PA-1
| Supramolecule | Name: Listeria monocytogenes man-PTS complexed with pediocin PA-1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Listeria monocytogenes (bacteria) Listeria monocytogenes (bacteria) |
-Macromolecule #1: Mannose/fructose/sorbose family PTS transporter subunit IIC
| Macromolecule | Name: Mannose/fructose/sorbose family PTS transporter subunit IIC type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Listeria monocytogenes (bacteria) Listeria monocytogenes (bacteria) |
| Molecular weight | Theoretical: 27.377561 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVISIILVV LIAFLAGIEG ILDEFQFHQP LIACTLIGLV TGNLTACIIL GGTLQMIALG WANIGAAVAP DAALASVASA IILVLGGQG VAGIPSAIAI AIPLAVAGLF LTMIVRTLAV PIVHLMDRAA EKGNIRSVEW LHISAICMQG IRIAIPAAAL L FIPADSVQ ...String: MSVISIILVV LIAFLAGIEG ILDEFQFHQP LIACTLIGLV TGNLTACIIL GGTLQMIALG WANIGAAVAP DAALASVASA IILVLGGQG VAGIPSAIAI AIPLAVAGLF LTMIVRTLAV PIVHLMDRAA EKGNIRSVEW LHISAICMQG IRIAIPAAAL L FIPADSVQ SFLEAMPAWL TDGMAIGGGM VVAVGYALVI NMMATKEVWP FFVIGFVVAA ISQLTLIAIG ALGVALALIY LN LSKMGGG NSNGGGGGNS RDPLGDILND Y UniProtKB: Mannose/fructose/sorbose family PTS transporter subunit IIC |
-Macromolecule #2: PTS mannose family transporter subunit IID
| Macromolecule | Name: PTS mannose family transporter subunit IID / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Listeria monocytogenes (bacteria) Listeria monocytogenes (bacteria) |
| Molecular weight | Theoretical: 33.402164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEKIELSKR DRLRVAWRST FIQGSWNYER MQNGGWAFSM IPAIKKLYKT KEDRSSALKR HLEFFNTHPY IASPILGVTL ALEEERANG AEVDDVAIQG VKVGMMGPLA GVGDPVFWFT IRPMLGALGA SLALSGNILG PILFFVAWNV IRWGFMWYTQ E FGYKAGSK ...String: MAEKIELSKR DRLRVAWRST FIQGSWNYER MQNGGWAFSM IPAIKKLYKT KEDRSSALKR HLEFFNTHPY IASPILGVTL ALEEERANG AEVDDVAIQG VKVGMMGPLA GVGDPVFWFT IRPMLGALGA SLALSGNILG PILFFVAWNV IRWGFMWYTQ E FGYKAGSK ITDDLSGGLL QDITKGASIL GMFVLAALVQ RWVNIQFAPI ISKVKLDEGA YIDWSHLPQG AQGIKTALQQ QQ AGLALSE IKVTTLQNNL DNLIPGLAAV ALTFLCMWLL KKKISPIIII LGLFVVGIVG HLIGLL UniProtKB: PTS mannose family transporter subunit IID |
-Macromolecule #3: Bacteriocin pediocin PA-1
| Macromolecule | Name: Bacteriocin pediocin PA-1 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pediococcus acidilactici (bacteria) Pediococcus acidilactici (bacteria) |
| Molecular weight | Theoretical: 5.001667 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PHMKYYGNGV TCGKHSCSVD WGKATTCIIN NGAMAWATGG HQGNHKC UniProtKB: Bacteriocin pediocin PA-1 |
-Macromolecule #4: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 4 / Number of copies: 3 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 30 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)