[English] 日本語
 Yorodumi
Yorodumi- EMDB-3048: Structure of a partial yeast 48S preinitiation complex in closed ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3048 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
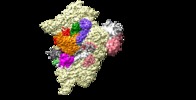
| Title | Structure of a partial yeast 48S preinitiation complex in closed conformation | |||||||||
 Map data Map data | To see a continuous density for factors eIF1, eIF2, mRNA, entire tRNA and eIF3 at the 40S subunit interface, apply a Gaussian filtering of 1.34 and a contour level of 0.025. To see a continuous density for the PCI domain of eIF3, apply a Gaussian filtering of 2.5 and a contour level of 0.008. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Eukaryotic translation initiation / 48S / small ribosome subunit. | |||||||||
| Function / homology |  Function and homology information Function and homology informationformation of translation initiation ternary complex / Recycling of eIF2:GDP / Cellular response to mitochondrial stress / eukaryotic translation initiation factor 3 complex, eIF3e / ABC-family proteins mediated transport / eukaryotic translation initiation factor 3 complex, eIF3m / methionyl-initiator methionine tRNA binding / incipient cellular bud site / translation reinitiation / eukaryotic translation initiation factor 2 complex ...formation of translation initiation ternary complex / Recycling of eIF2:GDP / Cellular response to mitochondrial stress / eukaryotic translation initiation factor 3 complex, eIF3e / ABC-family proteins mediated transport / eukaryotic translation initiation factor 3 complex, eIF3m / methionyl-initiator methionine tRNA binding / incipient cellular bud site / translation reinitiation / eukaryotic translation initiation factor 2 complex / eukaryotic translation initiation factor 3 complex / formation of cytoplasmic translation initiation complex / multi-eIF complex / cytoplasmic translational initiation / protein-synthesizing GTPase / eukaryotic 43S preinitiation complex / formation of translation preinitiation complex / eukaryotic 48S preinitiation complex / positive regulation of translational fidelity / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / Ribosomal scanning and start codon recognition / Formation of a pool of free 40S subunits / L13a-mediated translational silencing of Ceruloplasmin expression / ribosomal small subunit binding / 90S preribosome / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of translational fidelity / translation regulator activity / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / translation initiation factor binding / translation initiation factor activity / rescue of stalled ribosome / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / translational initiation / protein kinase C binding / maintenance of translational fidelity / modification-dependent protein catabolic process / cytoplasmic stress granule / protein tag activity / rRNA processing / ribosomal small subunit biogenesis / double-stranded RNA binding / small ribosomal subunit rRNA binding / ribosome binding / ribosomal small subunit assembly / small ribosomal subunit / cytosolic small ribosomal subunit / cytoplasmic translation / cytosolic large ribosomal subunit / rRNA binding / ribosome / structural constituent of ribosome / protein ubiquitination / translation / ribonucleoprotein complex / positive regulation of protein phosphorylation / GTPase activity / mRNA binding / ubiquitin protein ligase binding / nucleolus / GTP binding / protein kinase binding / RNA binding / zinc ion binding / identical protein binding / nucleus / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Kluyveromyces lactis (yeast) / Kluyveromyces lactis (yeast) /  | |||||||||
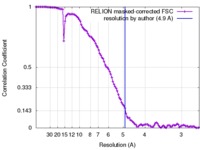
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Llacer JL / Hussain T / Ramakrishnan V | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2015 Journal: Mol Cell / Year: 2015Title: Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Authors: Jose L Llácer / Tanweer Hussain / Laura Marler / Colin Echeverría Aitken / Anil Thakur / Jon R Lorsch / Alan G Hinnebusch / V Ramakrishnan /   Abstract: Translation initiation in eukaryotes begins with the formation of a pre-initiation complex (PIC) containing the 40S ribosomal subunit, eIF1, eIF1A, eIF3, ternary complex (eIF2-GTP-Met-tRNAi), and ...Translation initiation in eukaryotes begins with the formation of a pre-initiation complex (PIC) containing the 40S ribosomal subunit, eIF1, eIF1A, eIF3, ternary complex (eIF2-GTP-Met-tRNAi), and eIF5. The PIC, in an open conformation, attaches to the 5' end of the mRNA and scans to locate the start codon, whereupon it closes to arrest scanning. We present single particle cryo-electron microscopy (cryo-EM) reconstructions of 48S PICs from yeast in these open and closed states, at 6.0 Å and 4.9 Å, respectively. These reconstructions show eIF2β as well as a configuration of eIF3 that appears to encircle the 40S, occupying part of the subunit interface. Comparison of the complexes reveals a large conformational change in the 40S head from an open mRNA latch conformation to a closed one that constricts the mRNA entry channel and narrows the P site to enclose tRNAi, thus elucidating key events in start codon recognition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3048.map.gz emd_3048.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3048-v30.xml emd-3048-v30.xml emd-3048.xml emd-3048.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3048_fsc.xml emd_3048_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  EMD-3048.png EMD-3048.png emd_3048.png emd_3048.png | 420.6 KB 420.6 KB | ||
| Others |  emd_3048_half_map_1.map.gz emd_3048_half_map_1.map.gz emd_3048_half_map_2.map.gz emd_3048_half_map_2.map.gz | 80.9 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3048 http://ftp.pdbj.org/pub/emdb/structures/EMD-3048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3048 | HTTPS FTP |
-Validation report
| Summary document |  emd_3048_validation.pdf.gz emd_3048_validation.pdf.gz | 437.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3048_full_validation.pdf.gz emd_3048_full_validation.pdf.gz | 436.7 KB | Display | |
| Data in XML |  emd_3048_validation.xml.gz emd_3048_validation.xml.gz | 11.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3048 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3048 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3048 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3048 | HTTPS FTP |
-Related structure data
| Related structure data |  3japMC  3047C  3049C  3050C  3jamC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3048.map.gz / Format: CCP4 / Size: 100.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3048.map.gz / Format: CCP4 / Size: 100.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | To see a continuous density for factors eIF1, eIF2, mRNA, entire tRNA and eIF3 at the 40S subunit interface, apply a Gaussian filtering of 1.34 and a contour level of 0.025. To see a continuous density for the PCI domain of eIF3, apply a Gaussian filtering of 2.5 and a contour level of 0.008. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
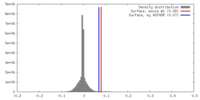
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 3048 half map 1.map
| File | emd_3048_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
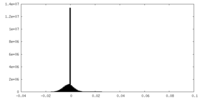
| Projections & Slices |
| ||||||||||||
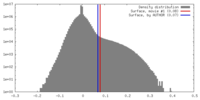
| Density Histograms |
-Supplemental map: emd 3048 half map 2.map
| File | emd_3048_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
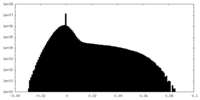
| Density Histograms |
- Sample components
Sample components
-Entire : Partial yeast 48S preinitiation complex
| Entire | Name: Partial yeast 48S preinitiation complex |
|---|---|
| Components |
|
-Supramolecule #1000: Partial yeast 48S preinitiation complex
| Supramolecule | Name: Partial yeast 48S preinitiation complex / type: sample / ID: 1000 / Oligomeric state: 1 / Number unique components: 7 |
|---|---|
| Molecular weight | Theoretical: 1.75 MDa |
-Supramolecule #1: Ribosome small subunit
| Supramolecule | Name: Ribosome small subunit / type: complex / ID: 1 / Name.synonym: 40S / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: SSU 40S, SSU RNA 18S |
|---|---|
| Source (natural) | Organism:  Kluyveromyces lactis (yeast) Kluyveromyces lactis (yeast) |
| Molecular weight | Theoretical: 1.2 MDa |
-Macromolecule #1: Eukaryotic initiation factor 1
| Macromolecule | Name: Eukaryotic initiation factor 1 / type: protein_or_peptide / ID: 1 / Name.synonym: eIF1 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.3 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Eukaryotic translation initiation factor eIF-1 |
-Macromolecule #2: Eukaryotic initiation factor 1A
| Macromolecule | Name: Eukaryotic initiation factor 1A / type: protein_or_peptide / ID: 2 / Name.synonym: eIF1A / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.4 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Eukaryotic translation initiation factor 1A |
-Macromolecule #3: Eukaryotic initiation factor 2
| Macromolecule | Name: Eukaryotic initiation factor 2 / type: protein_or_peptide / ID: 3 / Name.synonym: eIF2 Details: Uniprot codes are: alpha-P20459 beta-P09064 gamma-P32481 Number of copies: 1 / Oligomeric state: Three subunits, alpha, beta, gamma / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 124 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #4: Eukaryotic initiation factor 3
| Macromolecule | Name: Eukaryotic initiation factor 3 / type: protein_or_peptide / ID: 4 / Name.synonym: eIF3 Details: Uniprot codes are: 3a-P38249, 3b-P06103, 3c-P32497, 3g-A6ZZ25, 3i-P40217 Number of copies: 1 / Oligomeric state: Five subunits, 3a, 3b, 3c, 3g, 3i / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 395 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #5: Initiator transfer RNA
| Macromolecule | Name: Initiator transfer RNA / type: rna / ID: 5 / Name.synonym: Met-tRNAi Details: Doble mutation (G31U:C39A) when compared with yeast WT initiator tRNA. Classification: TRANSFER / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23 KDa |
| Sequence | String: AGCGCCGUGG CGCAGUGGAA GCGCGCAGGU CUCAUAAACC UGAUGUCCUC GGAUCGAAAC CGAGCGGCGC UACCA |
-Macromolecule #6: Messenger RNA
| Macromolecule | Name: Messenger RNA / type: rna / ID: 6 / Name.synonym: mRNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.4 KDa |
| Sequence | String: GGAAUCUCUC UCUAUGCUCU CUCUC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.17 mg/mL |
|---|---|
| Buffer | pH: 6.5 Details: 20mM MES-KOH, 40mM K-acetate, 10mM NH4-acetate, 8mM Mg-acetate, 2mM DTT |
| Grid | Details: Quantifoil R2/2 400 mesh copper grids with 4-5 nm thin carbon on top |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK I / Timed resolved state: 30 second incubation time / Method: Blot for 2.5-3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 59,000 times magnification |
| Details | Complete dataset was collected in 4 non-consecutive sessions |
| Date | Jul 4, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 5500 / Average electron dose: 27 e/Å2 Details: Complete dataset was collected in 4 non-consecutive sessions |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 104478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 78000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: 2 |
|---|---|
| Software | Name: Chimera, Coot, Refmac |
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Target criteria: R-factor, FSC |
| Output model |  PDB-3jap: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: K |
|---|---|
| Software | Name: Chimera, Coot, Refmac |
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Target criteria: R-factor, FSC |
| Output model |  PDB-3jap: |
-Atomic model buiding 3
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: C |
|---|---|
| Software | Name: Chimera, Coot, Refmac |
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Target criteria: R-factor, FSC |
| Output model |  PDB-3jap: |
-Atomic model buiding 4
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: Chimera, Coot, Refmac |
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Target criteria: R-factor, FSC |
| Output model |  PDB-3jap: |
-Atomic model buiding 5
| Initial model | PDB ID: Chain - #0 - Chain ID: B / Chain - #1 - Chain ID: I / Chain - #2 - Chain ID: G |
|---|---|
| Software | Name: Chimera, Coot, Refmac |
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Target criteria: R-factor, FSC |
| Output model |  PDB-3jap: |
 Movie
Movie Controller
Controller


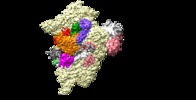

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)