[English] 日本語
 Yorodumi
Yorodumi- EMDB-27183: Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide i... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated wide(r) membrane tubes | ||||||||||||
 Map data Map data | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated wide(r) membrane tubes | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | nef / AP / trafficking / PROTEIN TRANSPORT | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationbasolateral protein secretion / negative regulation of CD4 production / perturbation by virus of host immune response / mitotic cleavage furrow ingression / endosome to melanosome transport / trans-Golgi Network Vesicle Budding / AP-1 adaptor complex / Lysosome Vesicle Biogenesis / protein trimerization / platelet dense granule organization ...basolateral protein secretion / negative regulation of CD4 production / perturbation by virus of host immune response / mitotic cleavage furrow ingression / endosome to melanosome transport / trans-Golgi Network Vesicle Budding / AP-1 adaptor complex / Lysosome Vesicle Biogenesis / protein trimerization / platelet dense granule organization / Glycosphingolipid transport / regulation of receptor internalization / melanosome assembly / Intra-Golgi traffic / symbiont-mediated suppression of host antigen processing and presentation of peptide antigen via MHC class I / regulation of Arp2/3 complex-mediated actin nucleation / Golgi to vacuole transport / symbiont-mediated suppression of host antigen processing and presentation of peptide antigen via MHC class II / Synthesis of PIPs at the Golgi membrane / Golgi Associated Vesicle Biogenesis / symbiont-mediated suppression of host apoptosis / clathrin adaptor activity / suppression by virus of host autophagy / MHC class II antigen presentation / CD4 receptor binding / Nef Mediated CD4 Down-regulation / thioesterase binding / dendritic spine organization / determination of left/right symmetry / long-term synaptic depression / clathrin-coated vesicle / COPI-dependent Golgi-to-ER retrograde traffic / positive regulation of memory T cell activation / Lysosome Vesicle Biogenesis / T cell mediated cytotoxicity directed against tumor cell target / TAP complex binding / positive regulation of CD8-positive, alpha-beta T cell activation / clathrin binding / CD8-positive, alpha-beta T cell activation / Golgi medial cisterna / Golgi Associated Vesicle Biogenesis / positive regulation of CD8-positive, alpha-beta T cell proliferation / CD8 receptor binding / cell leading edge / MHC class I protein binding / antigen processing and presentation of exogenous peptide antigen via MHC class I / Synthesis of PIPs at the plasma membrane / endoplasmic reticulum exit site / host cell Golgi membrane / antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent / TAP binding / protection from natural killer cell mediated cytotoxicity / intracellular copper ion homeostasis / beta-2-microglobulin binding / protein targeting / COPI-mediated anterograde transport / T cell receptor binding / detection of bacterium / clathrin-coated pit / vesicle-mediated transport / regulation of calcium-mediated signaling / viral life cycle / MHC class II antigen presentation / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / Neutrophil degranulation / sarcomere / kidney development / small monomeric GTPase / trans-Golgi network membrane / Nef mediated downregulation of MHC class I complex cell surface expression / antigen processing and presentation of endogenous peptide antigen via MHC class Ib / antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent / Endosomal/Vacuolar pathway / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / lumenal side of endoplasmic reticulum membrane / intracellular protein transport / virion component / trans-Golgi network / peptide antigen assembly with MHC class I protein complex / ER to Golgi transport vesicle membrane / cytoplasmic vesicle membrane / MHC class I peptide loading complex / T cell mediated cytotoxicity / positive regulation of T cell cytokine production / antigen processing and presentation of endogenous peptide antigen via MHC class I / cellular response to virus / MHC class I protein complex / SH3 domain binding / positive regulation of T cell mediated cytotoxicity / positive regulation of type II interferon production / recycling endosome membrane / phagocytic vesicle membrane / peptide antigen binding / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / Interferon gamma signaling / Interferon alpha/beta signaling / E3 ubiquitin ligases ubiquitinate target proteins / antibacterial humoral response / heart development / presynapse Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | ||||||||||||
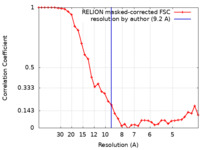
| Method | subtomogram averaging / cryo EM / Resolution: 9.2 Å | ||||||||||||
 Authors Authors | Hooy RM / Hurley JH | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat. Authors: Richard M Hooy / Yuichiro Iwamoto / Dan A Tudorica / Xuefeng Ren / James H Hurley /  Abstract: The adaptor protein (AP) complexes not only form the inner layer of clathrin coats but also have clathrin-independent roles in membrane traffic whose mechanisms are unknown. HIV-1 Nef hijacks AP-1 to ...The adaptor protein (AP) complexes not only form the inner layer of clathrin coats but also have clathrin-independent roles in membrane traffic whose mechanisms are unknown. HIV-1 Nef hijacks AP-1 to sequester major histocompatibility complex class I (MHC-I), evading immune detection. We found that AP-1:Arf1:Nef:MHC-I forms a coat on tubulated membranes without clathrin and determined its structure. The coat assembles via Arf1 dimer interfaces. AP-1-positive tubules are enriched in cells upon clathrin knockdown. Nef localizes preferentially to AP-1 tubules in cells, explaining how Nef sequesters MHC-I. Coat contact residues are conserved across Arf isoforms and the Arf-dependent AP complexes AP-1, AP-3, and AP-4. Thus, AP complexes can self-assemble with Arf1 into tubular coats without clathrin or other scaffolding factors. The AP-1:Arf1 coat defines the structural basis of a broader class of tubulovesicular membrane coats as an intermediate in clathrin vesicle formation from internal membranes and as an MHC-I sequestration mechanism in HIV-1 infection. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27183.map.gz emd_27183.map.gz | 2.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27183-v30.xml emd-27183-v30.xml emd-27183.xml emd-27183.xml | 30 KB 30 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27183_fsc.xml emd_27183_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27183.png emd_27183.png | 41.5 KB | ||
| Masks |  emd_27183_msk_1.map emd_27183_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27183.cif.gz emd-27183.cif.gz | 9 KB | ||
| Others |  emd_27183_half_map_1.map.gz emd_27183_half_map_1.map.gz emd_27183_half_map_2.map.gz emd_27183_half_map_2.map.gz | 3.1 MB 3.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27183 http://ftp.pdbj.org/pub/emdb/structures/EMD-27183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27183 | HTTPS FTP |
-Validation report
| Summary document |  emd_27183_validation.pdf.gz emd_27183_validation.pdf.gz | 783.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27183_full_validation.pdf.gz emd_27183_full_validation.pdf.gz | 783.1 KB | Display | |
| Data in XML |  emd_27183_validation.xml.gz emd_27183_validation.xml.gz | 9.4 KB | Display | |
| Data in CIF |  emd_27183_validation.cif.gz emd_27183_validation.cif.gz | 11.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27183 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27183 | HTTPS FTP |
-Related structure data
| Related structure data |  8d4eMC  7ux3C  8d4cC  8d4dC  8d4fC  8d4gC  8d9rC  8d9sC  8d9tC  8d9uC  8d9vC  8d9wC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27183.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27183.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated wide(r) membrane tubes | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27183_msk_1.map emd_27183_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_27183_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_27183_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of AP-1, Arf1, Nef and MHC-I cytosolic tail on a tubulate...
+Supramolecule #1: Complex of AP-1, Arf1, Nef and MHC-I cytosolic tail on a tubulate...
+Supramolecule #2: AP-1 heterotetramer
+Macromolecule #1: ADP-ribosylation factor 1
+Macromolecule #2: Protein Nef
+Macromolecule #3: HLA class I histocompatibility antigen, A alpha chain
+Macromolecule #4: AP-1 complex subunit beta-1
+Macromolecule #5: AP-1 complex subunit gamma-1
+Macromolecule #6: AP-1 complex subunit mu-1
+Macromolecule #7: AP-1 complex subunit sigma-3
+Macromolecule #8: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #9: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
Details: HEPES/KOAc concentrated stocks are diluted to their final concentrations then pH'd to 7.2 with KOH prior to use in experiments. | |||||||||||||||
| Grid | Model: EMS Lacey Carbon / Support film - Material: CARBON / Support film - topology: LACEY / Support film - Film thickness: 50 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: 60 second wait, 3-5 second blot, 597 filter paper, 0.5 second drain. Sample was supplemented with 10nm BSA-gold fiducials. 3.5ul of the mixture was double-side blotted.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 25 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 3.0 e/Å2 Details: Tilt images were collected in movie-mode. Each movie/tilt consisted of 3-4 frames each |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Protocol: RIGID BODY FIT | ||||||||
| Output model |  PDB-8d4e: |
 Movie
Movie Controller
Controller













































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)