[English] 日本語
 Yorodumi
Yorodumi- EMDB-25933: Yeast ATP synthase F1 region State 1catalytic(a) with 10 mM ATP -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast ATP synthase F1 region State 1catalytic(a) with 10 mM ATP | ||||||||||||
 Map data Map data | Sharpened map with Phenix density modification. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | F1-ATPase / ATP Synthase / Hydrolase / Nanomotor / Complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial protein degradation / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / mitochondrial nucleoid / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / mitochondrial intermembrane space ...Mitochondrial protein degradation / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / mitochondrial nucleoid / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / mitochondrial intermembrane space / mitochondrial inner membrane / mitochondrion / ATP binding / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
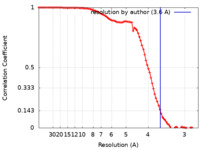
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Guo H / Rubinstein JL | ||||||||||||
| Funding support |  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of ATP synthase under strain during catalysis. Authors: Hui Guo / John L Rubinstein /  Abstract: ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor ...ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor subcomplex and are held stationary relative to each other by a peripheral stalk. Structures of resting mitochondrial ATP synthases revealed a left-handed curvature of the peripheral stalk even though rotation of the rotor, driven by either ATP hydrolysis in F or proton translocation through F, would apply a right-handed bending force to the stalk. We used cryoEM to image yeast mitochondrial ATP synthase under strain during ATP-hydrolysis-driven rotary catalysis, revealing a large deformation of the peripheral stalk. The structures show how the peripheral stalk opposes the bending force and suggests that during ATP synthesis proton translocation causes accumulation of strain in the stalk, which relaxes by driving the relative rotation of the rotor through six sub-steps within F, leading to catalysis. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25933.map.gz emd_25933.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25933-v30.xml emd-25933-v30.xml emd-25933.xml emd-25933.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25933_fsc.xml emd_25933_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_25933.png emd_25933.png | 87.8 KB | ||
| Masks |  emd_25933_msk_1.map emd_25933_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25933.cif.gz emd-25933.cif.gz | 6.8 KB | ||
| Others |  emd_25933_additional_1.map.gz emd_25933_additional_1.map.gz emd_25933_half_map_1.map.gz emd_25933_half_map_1.map.gz emd_25933_half_map_2.map.gz emd_25933_half_map_2.map.gz | 32.2 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25933 http://ftp.pdbj.org/pub/emdb/structures/EMD-25933 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25933 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25933 | HTTPS FTP |
-Related structure data
| Related structure data |  7tjvMC  7tjsC  7tjtC  7tjuC  7tjwC  7tjxC  7tjyC  7tjzC  7tk0C  7tk1C  7tk2C  7tk3C  7tk4C  7tk5C  7tk6C  7tk7C  7tk8C  7tk9C  7tkaC  7tkbC  7tkcC  7tkdC  7tkeC  7tkfC  7tkgC  7tkhC  7tkiC  7tkjC  7tkkC  7tklC  7tkmC  7tknC  7tkoC  7tkpC  7tkqC  7tkrC  7tksC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25933.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25933.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map with Phenix density modification. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3475 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25933_msk_1.map emd_25933_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map.
| File | emd_25933_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25933_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25933_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast ATP synthase F1 region State 1catalytic(a) with 10 mM ATP
| Entire | Name: Yeast ATP synthase F1 region State 1catalytic(a) with 10 mM ATP |
|---|---|
| Components |
|
-Supramolecule #1: Yeast ATP synthase F1 region State 1catalytic(a) with 10 mM ATP
| Supramolecule | Name: Yeast ATP synthase F1 region State 1catalytic(a) with 10 mM ATP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: ATP synthase subunit alpha
| Macromolecule | Name: ATP synthase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.007402 KDa |
| Sequence | String: ASTKAQPTEV SSILEERIKG VSDEANLNET GRVLAVGDGI ARVFGLNNIQ AEELVEFSSG VKGMALNLEP GQVGIVLFGS DRLVKEGEL VKRTGNIVDV PVGPGLLGRV VDALGNPIDG KGPIDAAGRS RAQVKAPGIL PRRSVHEPVQ TGLKAVDALV P IGRGQREL ...String: ASTKAQPTEV SSILEERIKG VSDEANLNET GRVLAVGDGI ARVFGLNNIQ AEELVEFSSG VKGMALNLEP GQVGIVLFGS DRLVKEGEL VKRTGNIVDV PVGPGLLGRV VDALGNPIDG KGPIDAAGRS RAQVKAPGIL PRRSVHEPVQ TGLKAVDALV P IGRGQREL IIGDRQTGKT AVALDTILNQ KRWNNGSDES KKLYCVYVAV GQKRSTVAQL VQTLEQHDAM KYSIIVAATA SE AAPLQYL APFTAASIGE WFRDNGKHAL IVYDDLSKQA VAYRQLSLLL RRPPGREAYP GDVFYLHSRL LERAAKLSEK EGS GSLTAL PVIETQGGDV SAYIPTNVIS ITDGQIFLEA ELFYKGIRPA INVGLSVSRV GSAAQVKALK QVAGSLKLFL AQYR EVAAF AQFGSDLDAS TKQTLVRGER LTQLLKQNQY SPLATEEQVP LIYAGVNGHL DGIELSRIGE FESSFLSYLK SNHNE LLTE IREKGELSKE LLASLKSATE SFVATF UniProtKB: ATP synthase subunit alpha, mitochondrial |
-Macromolecule #2: ATP synthase subunit beta
| Macromolecule | Name: ATP synthase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.181082 KDa |
| Sequence | String: ASAAQSTPIT GKVTAVIGAI VDVHFEQSEL PAILNALEIK TPQGKLVLEV AQHLGENTVR TIAMDGTEGL VRGEKVLDTG GPISVPVGR ETLGRIINVI GEPIDERGPI KSKLRKPIHA DPPSFAEQST SAEILETGIK VVDLLAPYAR GGKIGLFGGA G VGKTVFIQ ...String: ASAAQSTPIT GKVTAVIGAI VDVHFEQSEL PAILNALEIK TPQGKLVLEV AQHLGENTVR TIAMDGTEGL VRGEKVLDTG GPISVPVGR ETLGRIINVI GEPIDERGPI KSKLRKPIHA DPPSFAEQST SAEILETGIK VVDLLAPYAR GGKIGLFGGA G VGKTVFIQ ELINNIAKAH GGFSVFTGVG ERTREGNDLY REMKETGVIN LEGESKVALV FGQMNEPPGA RARVALTGLT IA EYFRDEE GQDVLLFIDN IFRFTQAGSE VSALLGRIPS AVGYQPTLAT DMGLLQERIT TTKKGSVTSV QAVYVPADDL TDP APATTF AHLDATTVLS RGISELGIYP AVDPLDSKSR LLDAAVVGQE HYDVASKVQE TLQTYKSLQD IIAILGMDEL SEQD KLTVE RARKIQRFLS QPFAVAEVFT GIPGKLVRLK DTVASFKAVL EGKYDNIPEH AFYMVGGIED VVAKAEKLAA EAN UniProtKB: ATP synthase subunit beta, mitochondrial |
-Macromolecule #3: ATP synthase subunit gamma
| Macromolecule | Name: ATP synthase subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.65716 KDa |
| Sequence | String: ATLKEVEMRL KSIKNIEKIT KTMKIVASTR LSKAEKAKIS AKKMDEAEQL FYKNAETKNL DVEATETGAP KELIVAITSD KGLCGSIHS QLAKAVRRHL NDQPNADIVT IGDKIKMQLL RTHPNNIKLS INGIGKDAPT FQESALIADK LLSVMKAGTY P KISIFYND ...String: ATLKEVEMRL KSIKNIEKIT KTMKIVASTR LSKAEKAKIS AKKMDEAEQL FYKNAETKNL DVEATETGAP KELIVAITSD KGLCGSIHS QLAKAVRRHL NDQPNADIVT IGDKIKMQLL RTHPNNIKLS INGIGKDAPT FQESALIADK LLSVMKAGTY P KISIFYND PVSSLSFEPS EKPIFNAKTI EQSPSFGKFE IDTDANVPRD LFEYTLANQM LTAMAQGYAA EISARRNAMD NA SKNAGDM INRYSILYNR TRQAVITNEL VDIITGASSL G UniProtKB: ATP synthase subunit gamma, mitochondrial |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 35 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 10037 / Average exposure time: 11.9 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 103896 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)