+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Yeast ATP synthase F1 region State 1catalytic(e-h) with 10 mM ATP | ||||||||||||
 マップデータ マップデータ | Sharpened map with Phenix density modification. | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | F1-ATPase / ATP Synthase / Hydrolase / Nanomotor / Complex | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Mitochondrial protein degradation / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / mitochondrial nucleoid / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / mitochondrial intermembrane space ...Mitochondrial protein degradation / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / mitochondrial nucleoid / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / mitochondrial intermembrane space / mitochondrial inner membrane / mitochondrion / ATP binding / cytosol 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  | ||||||||||||
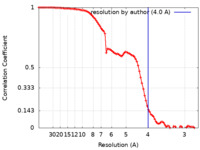
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.0 Å | ||||||||||||
 データ登録者 データ登録者 | Guo H / Rubinstein JL | ||||||||||||
| 資金援助 |  カナダ, 3件 カナダ, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2022 ジャーナル: Nat Commun / 年: 2022タイトル: Structure of ATP synthase under strain during catalysis. 著者: Hui Guo / John L Rubinstein /  要旨: ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor ...ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor subcomplex and are held stationary relative to each other by a peripheral stalk. Structures of resting mitochondrial ATP synthases revealed a left-handed curvature of the peripheral stalk even though rotation of the rotor, driven by either ATP hydrolysis in F or proton translocation through F, would apply a right-handed bending force to the stalk. We used cryoEM to image yeast mitochondrial ATP synthase under strain during ATP-hydrolysis-driven rotary catalysis, revealing a large deformation of the peripheral stalk. The structures show how the peripheral stalk opposes the bending force and suggests that during ATP synthesis proton translocation causes accumulation of strain in the stalk, which relaxes by driving the relative rotation of the rotor through six sub-steps within F, leading to catalysis. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_25934.map.gz emd_25934.map.gz | 59.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-25934-v30.xml emd-25934-v30.xml emd-25934.xml emd-25934.xml | 23.2 KB 23.2 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_25934_fsc.xml emd_25934_fsc.xml | 8.8 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_25934.png emd_25934.png | 83.8 KB | ||
| マスクデータ |  emd_25934_msk_1.map emd_25934_msk_1.map | 64 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-25934.cif.gz emd-25934.cif.gz | 6.7 KB | ||
| その他 |  emd_25934_additional_1.map.gz emd_25934_additional_1.map.gz emd_25934_half_map_1.map.gz emd_25934_half_map_1.map.gz emd_25934_half_map_2.map.gz emd_25934_half_map_2.map.gz | 32.2 MB 59.5 MB 59.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25934 http://ftp.pdbj.org/pub/emdb/structures/EMD-25934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25934 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7tjwMC  7tjsC  7tjtC  7tjuC  7tjvC  7tjxC  7tjyC  7tjzC  7tk0C  7tk1C  7tk2C  7tk3C  7tk4C  7tk5C  7tk6C  7tk7C  7tk8C  7tk9C  7tkaC  7tkbC  7tkcC  7tkdC  7tkeC  7tkfC  7tkgC  7tkhC  7tkiC  7tkjC  7tkkC  7tklC  7tkmC  7tknC  7tkoC  7tkpC  7tkqC  7tkrC  7tksC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_25934.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_25934.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Sharpened map with Phenix density modification. | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.3475 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_25934_msk_1.map emd_25934_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: Unsharpened map.
| ファイル | emd_25934_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Unsharpened map. | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_25934_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_25934_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Yeast ATP synthase F1 region State 1catalytic(e-h) with 10 mM ATP
| 全体 | 名称: Yeast ATP synthase F1 region State 1catalytic(e-h) with 10 mM ATP |
|---|---|
| 要素 |
|
-超分子 #1: Yeast ATP synthase F1 region State 1catalytic(e-h) with 10 mM ATP
| 超分子 | 名称: Yeast ATP synthase F1 region State 1catalytic(e-h) with 10 mM ATP タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#3 |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 400 KDa |
-分子 #1: ATP synthase subunit alpha
| 分子 | 名称: ATP synthase subunit alpha / タイプ: protein_or_peptide / ID: 1 / コピー数: 3 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 55.007402 KDa |
| 配列 | 文字列: ASTKAQPTEV SSILEERIKG VSDEANLNET GRVLAVGDGI ARVFGLNNIQ AEELVEFSSG VKGMALNLEP GQVGIVLFGS DRLVKEGEL VKRTGNIVDV PVGPGLLGRV VDALGNPIDG KGPIDAAGRS RAQVKAPGIL PRRSVHEPVQ TGLKAVDALV P IGRGQREL ...文字列: ASTKAQPTEV SSILEERIKG VSDEANLNET GRVLAVGDGI ARVFGLNNIQ AEELVEFSSG VKGMALNLEP GQVGIVLFGS DRLVKEGEL VKRTGNIVDV PVGPGLLGRV VDALGNPIDG KGPIDAAGRS RAQVKAPGIL PRRSVHEPVQ TGLKAVDALV P IGRGQREL IIGDRQTGKT AVALDTILNQ KRWNNGSDES KKLYCVYVAV GQKRSTVAQL VQTLEQHDAM KYSIIVAATA SE AAPLQYL APFTAASIGE WFRDNGKHAL IVYDDLSKQA VAYRQLSLLL RRPPGREAYP GDVFYLHSRL LERAAKLSEK EGS GSLTAL PVIETQGGDV SAYIPTNVIS ITDGQIFLEA ELFYKGIRPA INVGLSVSRV GSAAQVKALK QVAGSLKLFL AQYR EVAAF AQFGSDLDAS TKQTLVRGER LTQLLKQNQY SPLATEEQVP LIYAGVNGHL DGIELSRIGE FESSFLSYLK SNHNE LLTE IREKGELSKE LLASLKSATE SFVATF UniProtKB: ATP synthase subunit alpha, mitochondrial |
-分子 #2: ATP synthase subunit beta
| 分子 | 名称: ATP synthase subunit beta / タイプ: protein_or_peptide / ID: 2 / コピー数: 3 / 光学異性体: LEVO / EC番号: H+-transporting two-sector ATPase |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 51.181082 KDa |
| 配列 | 文字列: ASAAQSTPIT GKVTAVIGAI VDVHFEQSEL PAILNALEIK TPQGKLVLEV AQHLGENTVR TIAMDGTEGL VRGEKVLDTG GPISVPVGR ETLGRIINVI GEPIDERGPI KSKLRKPIHA DPPSFAEQST SAEILETGIK VVDLLAPYAR GGKIGLFGGA G VGKTVFIQ ...文字列: ASAAQSTPIT GKVTAVIGAI VDVHFEQSEL PAILNALEIK TPQGKLVLEV AQHLGENTVR TIAMDGTEGL VRGEKVLDTG GPISVPVGR ETLGRIINVI GEPIDERGPI KSKLRKPIHA DPPSFAEQST SAEILETGIK VVDLLAPYAR GGKIGLFGGA G VGKTVFIQ ELINNIAKAH GGFSVFTGVG ERTREGNDLY REMKETGVIN LEGESKVALV FGQMNEPPGA RARVALTGLT IA EYFRDEE GQDVLLFIDN IFRFTQAGSE VSALLGRIPS AVGYQPTLAT DMGLLQERIT TTKKGSVTSV QAVYVPADDL TDP APATTF AHLDATTVLS RGISELGIYP AVDPLDSKSR LLDAAVVGQE HYDVASKVQE TLQTYKSLQD IIAILGMDEL SEQD KLTVE RARKIQRFLS QPFAVAEVFT GIPGKLVRLK DTVASFKAVL EGKYDNIPEH AFYMVGGIED VVAKAEKLAA EAN UniProtKB: ATP synthase subunit beta, mitochondrial |
-分子 #3: ATP synthase subunit gamma
| 分子 | 名称: ATP synthase subunit gamma / タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 30.65716 KDa |
| 配列 | 文字列: ATLKEVEMRL KSIKNIEKIT KTMKIVASTR LSKAEKAKIS AKKMDEAEQL FYKNAETKNL DVEATETGAP KELIVAITSD KGLCGSIHS QLAKAVRRHL NDQPNADIVT IGDKIKMQLL RTHPNNIKLS INGIGKDAPT FQESALIADK LLSVMKAGTY P KISIFYND ...文字列: ATLKEVEMRL KSIKNIEKIT KTMKIVASTR LSKAEKAKIS AKKMDEAEQL FYKNAETKNL DVEATETGAP KELIVAITSD KGLCGSIHS QLAKAVRRHL NDQPNADIVT IGDKIKMQLL RTHPNNIKLS INGIGKDAPT FQESALIADK LLSVMKAGTY P KISIFYND PVSSLSFEPS EKPIFNAKTI EQSPSFGKFE IDTDANVPRD LFEYTLANQM LTAMAQGYAA EISARRNAMD NA SKNAGDM INRYSILYNR TRQAVITNEL VDIITGASSL G UniProtKB: ATP synthase subunit gamma, mitochondrial |
-分子 #4: ADENOSINE-5'-TRIPHOSPHATE
| 分子 | 名称: ADENOSINE-5'-TRIPHOSPHATE / タイプ: ligand / ID: 4 / コピー数: 5 / 式: ATP |
|---|---|
| 分子量 | 理論値: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-分子 #5: MAGNESIUM ION
| 分子 | 名称: MAGNESIUM ION / タイプ: ligand / ID: 5 / コピー数: 5 / 式: MG |
|---|---|
| 分子量 | 理論値: 24.305 Da |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 15 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 |
| グリッド | モデル: Homemade / 材質: COPPER/RHODIUM / メッシュ: 300 / 支持フィルム - 材質: GOLD / 支持フィルム - トポロジー: HOLEY / 支持フィルム - Film thickness: 35 / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 120 sec. |
| 凍結 | 凍結剤: ETHANE-PROPANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: LEICA EM GP |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON IV (4k x 4k) デジタル化 - サイズ - 横: 4096 pixel / デジタル化 - サイズ - 縦: 4096 pixel / 実像数: 10037 / 平均露光時間: 11.9 sec. / 平均電子線量: 40.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 50.0 µm / 倍率(補正後): 103896 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 1.1 µm / 倍率(公称値): 59000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー

























































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)