[English] 日本語
 Yorodumi
Yorodumi- EMDB-23883: Single-Particle Cryo-EM Structure of Major Facilitator Superfamil... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23883 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
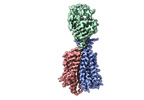
| Title | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | ||||||||||||||||||||||||
 Map data Map data | MFSD2A_gallus_gallus_main_map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationSynthesis of PC / fatty acid transmembrane transporter activity / lysophospholipid translocation / lysophospholipid:sodium symporter activity / lipid transport across blood-brain barrier / carbohydrate transport / fatty acid transport / endoplasmic reticulum membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
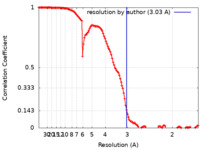
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | ||||||||||||||||||||||||
 Authors Authors | Cater RJ / Chua GL / Erramilli SK / Keener JE / Choy BC / Tokarz P / Chin CF / Quek DQY / Kloss B / Pepe JG ...Cater RJ / Chua GL / Erramilli SK / Keener JE / Choy BC / Tokarz P / Chin CF / Quek DQY / Kloss B / Pepe JG / Parisi G / Wong BH / Clarke OB / Marty MT / Kossiakoff AA / Khelashvili G / Silver DL / Mancia F | ||||||||||||||||||||||||
| Funding support |  United States, United States,  Singapore, 7 items Singapore, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis of omega-3 fatty acid transport across the blood-brain barrier. Authors: Rosemary J Cater / Geok Lin Chua / Satchal K Erramilli / James E Keener / Brendon C Choy / Piotr Tokarz / Cheen Fei Chin / Debra Q Y Quek / Brian Kloss / Joseph G Pepe / Giacomo Parisi / ...Authors: Rosemary J Cater / Geok Lin Chua / Satchal K Erramilli / James E Keener / Brendon C Choy / Piotr Tokarz / Cheen Fei Chin / Debra Q Y Quek / Brian Kloss / Joseph G Pepe / Giacomo Parisi / Bernice H Wong / Oliver B Clarke / Michael T Marty / Anthony A Kossiakoff / George Khelashvili / David L Silver / Filippo Mancia /   Abstract: Docosahexaenoic acid is an omega-3 fatty acid that is essential for neurological development and function, and it is supplied to the brain and eyes predominantly from dietary sources. This nutrient ...Docosahexaenoic acid is an omega-3 fatty acid that is essential for neurological development and function, and it is supplied to the brain and eyes predominantly from dietary sources. This nutrient is transported across the blood-brain and blood-retina barriers in the form of lysophosphatidylcholine by major facilitator superfamily domain containing 2A (MFSD2A) in a Na-dependent manner. Here we present the structure of MFSD2A determined using single-particle cryo-electron microscopy, which reveals twelve transmembrane helices that are separated into two pseudosymmetric domains. The transporter is in an inward-facing conformation and features a large amphipathic cavity that contains the Na-binding site and a bound lysolipid substrate, which we confirmed using native mass spectrometry. Together with our functional analyses and molecular dynamics simulations, this structure reveals details of how MFSD2A interacts with substrates and how Na-dependent conformational changes allow for the release of these substrates into the membrane through a lateral gate. Our work provides insights into the molecular mechanism by which this atypical major facility superfamily transporter mediates the uptake of lysolipids into the brain, and has the potential to aid in the delivery of neurotherapeutic agents. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23883.map.gz emd_23883.map.gz | 168.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23883-v30.xml emd-23883-v30.xml emd-23883.xml emd-23883.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23883_fsc.xml emd_23883_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_23883.png emd_23883.png | 55.1 KB | ||
| Masks |  emd_23883_msk_1.map emd_23883_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_23883_half_map_1.map.gz emd_23883_half_map_1.map.gz emd_23883_half_map_2.map.gz emd_23883_half_map_2.map.gz | 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23883 http://ftp.pdbj.org/pub/emdb/structures/EMD-23883 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23883 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23883 | HTTPS FTP |
-Validation report
| Summary document |  emd_23883_validation.pdf.gz emd_23883_validation.pdf.gz | 1021.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23883_full_validation.pdf.gz emd_23883_full_validation.pdf.gz | 1020.6 KB | Display | |
| Data in XML |  emd_23883_validation.xml.gz emd_23883_validation.xml.gz | 20.2 KB | Display | |
| Data in CIF |  emd_23883_validation.cif.gz emd_23883_validation.cif.gz | 25.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23883 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23883 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23883 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23883 | HTTPS FTP |
-Related structure data
| Related structure data |  7mjsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10698 (Title: Structural basis of ω-3 fatty acid transport across the blood-brain barrier EMPIAR-10698 (Title: Structural basis of ω-3 fatty acid transport across the blood-brain barrierData size: 3.9 TB Data #1: MFSD2A_GG_compled with 2AG3 Fab and LPC-18:3 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23883.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23883.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MFSD2A_gallus_gallus_main_map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
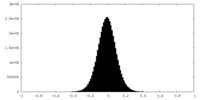
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23883_msk_1.map emd_23883_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
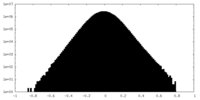
| Density Histograms |
-Half map: #1
| File | emd_23883_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_23883_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Single-Particle Cryo-EM Structure of Major Facilitator Superfamil...
| Entire | Name: Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A (MFSD2A) Gallus gallus in complex with LPC-18:3 |
|---|---|
| Components |
|
-Supramolecule #1: Single-Particle Cryo-EM Structure of Major Facilitator Superfamil...
| Supramolecule | Name: Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A (MFSD2A) Gallus gallus in complex with LPC-18:3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 2AG3 Fab heavy chain
| Macromolecule | Name: 2AG3 Fab heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) |
| Molecular weight | Theoretical: 25.646537 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NIYSSSIHWV RQAPGKGLEW VAYIYSYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSLEYLYSS GYQYKWATGL DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NIYSSSIHWV RQAPGKGLEW VAYIYSYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSLEYLYSS GYQYKWATGL DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDKT HT |
-Macromolecule #2: 2AG3 Fab light chain
| Macromolecule | Name: 2AG3 Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) |
| Molecular weight | Theoretical: 23.655182 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSFYNQPF TFGQGTKVEI KRTVAAPSVF IFPPSDSQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS G NSQESVTE ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSFYNQPF TFGQGTKVEI KRTVAAPSVF IFPPSDSQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS G NSQESVTE QDSKDSTYSL SSTLTLSKAD YEKHKVYACE VTHQGLSSPV TKSFNRGEC |
-Macromolecule #3: Major Facilitator Superfamily Domain containing 2A
| Macromolecule | Name: Major Facilitator Superfamily Domain containing 2A / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.789168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGGGGAERV RVGAAAAGLL PPSCRQPRRR ESRERLSVCS KLCYAVGGAP YQTTGCALGF FLQIYLLDVA QLDPFYASII LFVGRAWDA ITDPMVGFFI SKTPWTRFGR LMPWIIFSTP FAVISYFLIW FVPDISTGQV MWYLIFYCIF QTLVTCFHVP Y SALTMFIS ...String: MAGGGGAERV RVGAAAAGLL PPSCRQPRRR ESRERLSVCS KLCYAVGGAP YQTTGCALGF FLQIYLLDVA QLDPFYASII LFVGRAWDA ITDPMVGFFI SKTPWTRFGR LMPWIIFSTP FAVISYFLIW FVPDISTGQV MWYLIFYCIF QTLVTCFHVP Y SALTMFIS REQSERDSAT AYRMTVEVLG TVLGTAIQGQ IVGKAVTPCI ENPPFLSETN FSVAIRNVNM THYTGSLADT RN AYMVAAG VIGGLYILCA VILSVGVREK RESSELQSDE PVSFFRGLKL VMNHGAYIKL ITGFLFTSLA FMLLEGNFAL FCT YTLGFR NEFQNILLAI MLSATLTIPF WQWFLTRFGK KTAVYVGISS AVPFLITVVV LDSNLVVTYI VAVAAGISVA AAFL LPWSM LPDVIDDFKL QHPESRGHEA IFFSFYVFFT KFTSGVSLGI STLSLDFAGY QTRGCSQPSE VNITLKLLVS AVPVG LILL GLLLFKLYPI DEEKRRENKK ALQDLREESN SSSESDSTEL ANIVENLYFQ GSHHHHHHHH HH |
-Macromolecule #4: [(2~{R})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethox...
| Macromolecule | Name: [(2~{R})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propyl] (9~{Z},12~{Z},15~{Z})-octadeca-9,12,15-trienoate type: ligand / ID: 4 / Number of copies: 1 / Formula: ZGS |
|---|---|
| Molecular weight | Theoretical: 518.644 Da |
| Chemical component information | 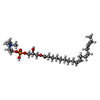 ChemComp-ZGS: |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Digitization - Sampling interval: 5.0 µm / Average exposure time: 3.0 sec. / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)