+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22855 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Partial Cas6-RT-Cas1--Cas2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Hydrolase / CRISPR Cas Protein / Reverse Transcriptase | |||||||||
| Biological species |  Thiomicrospira sp. (bacteria) Thiomicrospira sp. (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Hoel CM / Wang JY | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex. Authors: Joy Y Wang / Christopher M Hoel / Basem Al-Shayeb / Jillian F Banfield / Stephen G Brohawn / Jennifer A Doudna /  Abstract: CRISPR-Cas systems provide adaptive immunity in bacteria and archaea, beginning with integration of foreign sequences into the host CRISPR genomic locus and followed by transcription and maturation ...CRISPR-Cas systems provide adaptive immunity in bacteria and archaea, beginning with integration of foreign sequences into the host CRISPR genomic locus and followed by transcription and maturation of CRISPR RNAs (crRNAs). In some CRISPR systems, a reverse transcriptase (RT) fusion to the Cas1 integrase and Cas6 maturase creates a single protein that enables concerted sequence integration and crRNA production. To elucidate how the RT-integrase organizes distinct enzymatic activities, we present the cryo-EM structure of a Cas6-RT-Cas1-Cas2 CRISPR integrase complex. The structure reveals a heterohexamer in which the RT directly contacts the integrase and maturase domains, suggesting functional coordination between all three active sites. Together with biochemical experiments, our data support a model of sequential enzymatic activities that enable CRISPR sequence acquisition from RNA and DNA substrates. These findings highlight an expanded capacity of some CRISPR systems to acquire diverse sequences that direct CRISPR-mediated interference. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22855.map.gz emd_22855.map.gz | 225.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22855-v30.xml emd-22855-v30.xml emd-22855.xml emd-22855.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22855.png emd_22855.png | 143.7 KB | ||
| Filedesc metadata |  emd-22855.cif.gz emd-22855.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22855 http://ftp.pdbj.org/pub/emdb/structures/EMD-22855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22855 | HTTPS FTP |
-Related structure data
| Related structure data |  7kftMC  7kfuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10642 (Title: Cas6-reverse transcriptase-Cas1—Cas2 CRISPR integrase complex EMPIAR-10642 (Title: Cas6-reverse transcriptase-Cas1—Cas2 CRISPR integrase complexData size: 2.2 TB Data #1: Unaligned multi-frame movies of a Cas6-reverse transcriptase-Cas1—Cas2 CRISPR integrase complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22855.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22855.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.187 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
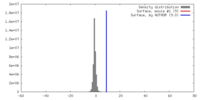
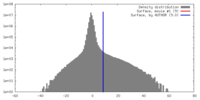
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cas6-RT-Cas1--Cas2 complex
| Entire | Name: Cas6-RT-Cas1--Cas2 complex |
|---|---|
| Components |
|
-Supramolecule #1: Cas6-RT-Cas1--Cas2 complex
| Supramolecule | Name: Cas6-RT-Cas1--Cas2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Thiomicrospira sp. (bacteria) Thiomicrospira sp. (bacteria) |
| Molecular weight | Theoretical: 470 KDa |
-Macromolecule #1: Cas2
| Macromolecule | Name: Cas2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thiomicrospira sp. (bacteria) Thiomicrospira sp. (bacteria) |
| Molecular weight | Theoretical: 11.583263 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMKHYLIC FDVQHDKTRA KLSRLLEKYG PRVQGSVFEV SFKTPDRKRQ LEYKIHQIIK QSNTEENNIR FYNLNKDTIK HSHDINGNP IAQLPAAIVL |
-Macromolecule #2: Cas6-RT-Cas1
| Macromolecule | Name: Cas6-RT-Cas1 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thiomicrospira sp. (bacteria) Thiomicrospira sp. (bacteria) |
| Molecular weight | Theoretical: 112.869414 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMILPSFP DLTGLVVNLK FTARAEFSLN HEMAVDAFLR HSLNLGESYS HHLSIITPEN GRLFYREGDT YRFVVIAMGN QQQTNSIWH TLINHLRKLP DSAPITDKQA PLRNNIKLES LNDLFDGIPV SSKESLDAYT LQRAMEQGLA WHKAANLTEQ P LDIQWYWQ ...String: SNAMILPSFP DLTGLVVNLK FTARAEFSLN HEMAVDAFLR HSLNLGESYS HHLSIITPEN GRLFYREGDT YRFVVIAMGN QQQTNSIWH TLINHLRKLP DSAPITDKQA PLRNNIKLES LNDLFDGIPV SSKESLDAYT LQRAMEQGLA WHKAANLTEQ P LDIQWYWQ STVRILHADH KQHKGEQRYC RDAVQLTPLL LLKRIYETLN NVATYFGLKT NKNTTENHQA WLKEQAQYIE IQ HPDLYWI DTPYFGKDAE KNTLGGMAGN FTLSLKPGIE PGLLAMLILT QMVGVGQRRT SGLGKYWLKH SLKHAHLILG LKP NRVTRS QTLLDCIIQP HIISQAIAEI EKKTNIDTLN ERTLSQVQSA IGQLRKHQYQ APKLQGFTIP KKDGTERLLA VSPL YDRIL QKAAAIVLTP GLDAIMSQAS YGYRKGLSRQ QVRYEIQNAY RQGYHWVYES DIEDFFDAVY RPQLINRLKS LLGND PLWE QIESWLGQDI HIKDTIIERT PNLGLPQGSP LSPLLANFIL DDFDSDLETH GFKIIRFADD FIILCKSQHE AQQAAH AVE QSLKEVKLSI NVEKTHIIQL NQGFRFLGYL FREDHAIEIA GEKSDGRTTF AAEQTPTNLP PWLANLGTKS PQPLADD DL PKKSYGQIET QGTHLVLAGD AQIITTDNQN LIVKKDDKIT HKISLEQLHA VTLIGLHTMT LPAKHRLLEH KIPVHIAD R TGRYLGAVTS FQPAQNNYKN WFIQLQMCDR EPFAHAIAQQ IVISRIHNQR QTLLKRKAHR KQLQQTLSNL KKLQYKVTA ATKRSSLNGL EGSATREYFQ QFNLFLPEWA HFSKRTRRPP KDPFNVLLSL GYTILYSHTD AILQSAGFIT WKGIYHQQSA AHAALASDI MESYRHLVER YAIYIINHGQ IKQDDFRQEK DHLGQDTIRL SAEARRRYVG GLINRFQKFS KDKTLHQHLY Q QAQQLKNA MHNQQSSQFQ VWKELK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 1, 3 second blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: cryoSPARC ab initio |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 129175 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 3.0) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 3.1) |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)