[English] 日本語
 Yorodumi
Yorodumi- EMDB-21429: Density-fitted Model Structure of Antibody Variable Domains of Ty... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21429 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Density-fitted Model Structure of Antibody Variable Domains of TyTx11 in Complex with Typhoid Toxin | ||||||||||||
 Map data Map data | Full map | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationTransferases; Glycosyltransferases; Pentosyltransferases / : / NAD+ ADP-ribosyltransferase activity / toxin activity / endonuclease activity / Hydrolases; Acting on ester bonds / hydrolase activity / extracellular region Similarity search - Function | ||||||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) / Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) /  | ||||||||||||
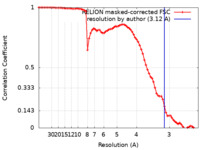
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | ||||||||||||
 Authors Authors | Nguyen T / Song J | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: iScience / Year: 2021 Journal: iScience / Year: 2021Title: Mechanisms of typhoid toxin neutralization by antibodies targeting glycan receptor binding and nuclease subunits. Authors: Changhwan Ahn / Yi-An Yang / Durga P Neupane / Tri Nguyen / Angelene F Richards / Ji Hyun Sim / Nicholas J Mantis / Jeongmin Song /  Abstract: Nearly all clinical isolates of Typhi, the cause of typhoid fever, are antibiotic resistant. All Typhi isolates secrete an AB exotoxin called typhoid toxin to benefit the pathogen during infection. ...Nearly all clinical isolates of Typhi, the cause of typhoid fever, are antibiotic resistant. All Typhi isolates secrete an AB exotoxin called typhoid toxin to benefit the pathogen during infection. Here, we demonstrate that antibiotic-resistant Typhi secretes typhoid toxin continuously during infection regardless of antibiotic treatment. We characterize typhoid toxin antibodies targeting glycan-receptor-binding PltB or nuclease CdtB, which neutralize typhoid toxin and , as demonstrated by using typhoid toxin secreted by antibiotic-resistant Typhi during human cell infection and lethal dose typhoid toxin challenge to mice. TyTx11 generated in this study neutralizes typhoid toxin effectively, comparable to TyTx4 that binds to all PltB subunits available per holotoxin. Cryoelectron microscopy explains that the binding of TyTx11 to CdtB makes this subunit inactive through CdtB catalytic-site conformational change. The identified toxin-neutralizing epitopes are conserved across all Typhi clinical isolates, offering critical insights into typhoid toxin-neutralizing strategies. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21429.map.gz emd_21429.map.gz | 26.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21429-v30.xml emd-21429-v30.xml emd-21429.xml emd-21429.xml | 28.4 KB 28.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21429_fsc.xml emd_21429_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_21429.png emd_21429.png | 54.1 KB | ||
| Masks |  emd_21429_msk_1.map emd_21429_msk_1.map | 34.3 MB |  Mask map Mask map | |
| Others |  emd_21429_additional_1.map.gz emd_21429_additional_1.map.gz emd_21429_additional_2.map.gz emd_21429_additional_2.map.gz emd_21429_half_map_1.map.gz emd_21429_half_map_1.map.gz emd_21429_half_map_2.map.gz emd_21429_half_map_2.map.gz | 32.2 MB 2.5 MB 26.4 MB 26.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21429 http://ftp.pdbj.org/pub/emdb/structures/EMD-21429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21429 | HTTPS FTP |
-Validation report
| Summary document |  emd_21429_validation.pdf.gz emd_21429_validation.pdf.gz | 188.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21429_full_validation.pdf.gz emd_21429_full_validation.pdf.gz | 188.4 KB | Display | |
| Data in XML |  emd_21429_validation.xml.gz emd_21429_validation.xml.gz | 502 B | Display | |
| Data in CIF |  emd_21429_validation.cif.gz emd_21429_validation.cif.gz | 373 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21429 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21429 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21429 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21429 | HTTPS FTP |
-Related structure data
| Related structure data |  6vx4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21429.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21429.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
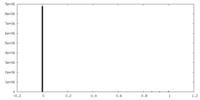
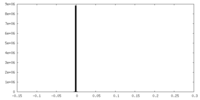
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21429_msk_1.map emd_21429_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
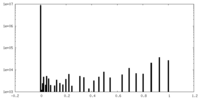
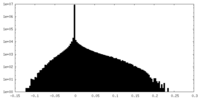
| Density Histograms |
-Additional map: RELION 3 Postprocess Map
| File | emd_21429_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION 3 Postprocess Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
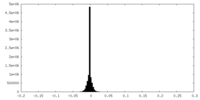
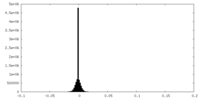
| Density Histograms |
-Additional map: RELION 3 Postprocess Map - Masked
| File | emd_21429_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION 3 Postprocess Map - Masked | ||||||||||||
| Projections & Slices |
| ||||||||||||
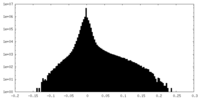
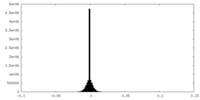
| Density Histograms |
-Half map: Half map 1
| File | emd_21429_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_21429_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of TyTx11 Fab with Typhoid toxin
| Entire | Name: Complex of TyTx11 Fab with Typhoid toxin |
|---|---|
| Components |
|
-Supramolecule #1: Complex of TyTx11 Fab with Typhoid toxin
| Supramolecule | Name: Complex of TyTx11 Fab with Typhoid toxin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Map of Fab segment of IgG antibody TyTx11 in complex with purified Typhoid toxin |
|---|
-Supramolecule #2: Typhoid toxin
| Supramolecule | Name: Typhoid toxin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2, #4-#5 / Details: S. Typhi A2B5 toxin wild type |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) |
| Recombinant expression | Organism:  |
-Supramolecule #3: TyTx11 Fab
| Supramolecule | Name: TyTx11 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1, #3 / Details: Fab segment of IgG antibody TyTx11 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Variable Domain of Kappa Chain of TyTx11 Antibody
| Macromolecule | Name: Variable Domain of Kappa Chain of TyTx11 Antibody / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.689935 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSSSY LSVSQGDRVT ITCKASDHID NWLAWYQQKP GNAPRLLISG ATSLKTGLPS RFSGSGSGKD FSLSITNLQT EDVASYYCQ QYWRTPYTFG GGTKLEI |
-Macromolecule #2: Pertussis like toxin subunit B
| Macromolecule | Name: Pertussis like toxin subunit B / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) |
| Molecular weight | Theoretical: 12.563042 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EWTGDNTNAY YSDEVISELH VGQIDTSPYF CIKTVKANGS GTPVVACAVS KQSIWAPSFK ELLDQARYFY STGQSVRIHV QKNIWTYPL FVNTFSANAL VGLSSCSATQ CFGPK |
-Macromolecule #3: Variable Domain of Heavy Chain of Antibody TyTx11
| Macromolecule | Name: Variable Domain of Heavy Chain of Antibody TyTx11 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.049723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLQQSGPV LVKPGPSVKI SCKASGYSFI DYFMNWVMQS HGKSLEWIGR IDPYSGDTFY NQKFKGKATL TVDKSSTTAH MELRSLASE DSAVYYCARE VLNYYAMDYW GQGTSVTVSS AKTTPPSVYP LAPGSAAQTN SMVTLGCLVK GYFPEPVTVT W NSGSPWYI SFLKLNKIF(UNK) RE(UNK) |
-Macromolecule #4: Pertussis toxin-like subunit ArtA
| Macromolecule | Name: Pertussis toxin-like subunit ArtA / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) |
| Molecular weight | Theoretical: 25.117145 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VDFVYRVDST PPDVIFRDGF SLLGYNRNFQ QFISGRSCSG GSSDSRYIAT TSSVNQTYAI ARAYYSRSTF KGNLYRYQIR ADNNFYSLL PSITYLETQG GHFNAYEKTM MRLQREYVST LSILPENIQK AVALVYDSAT GLVKDGVSTM NASYLGLSTT S NPGVIPFL ...String: VDFVYRVDST PPDVIFRDGF SLLGYNRNFQ QFISGRSCSG GSSDSRYIAT TSSVNQTYAI ARAYYSRSTF KGNLYRYQIR ADNNFYSLL PSITYLETQG GHFNAYEKTM MRLQREYVST LSILPENIQK AVALVYDSAT GLVKDGVSTM NASYLGLSTT S NPGVIPFL PEPQTYTQQR IDAFGPLISS CFSIGSVCHS HRGQRADVYN MSFYDARPVI ELILSK |
-Macromolecule #5: Cytolethal distending toxin subunit B
| Macromolecule | Name: Cytolethal distending toxin subunit B / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) Salmonella enterica subsp. enterica serovar Typhi str. CT18 (bacteria) |
| Molecular weight | Theoretical: 28.149777 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NISDYKVMTW NLQGSSASTE SKWNVNVRQL LSGTAGVDIL MVQEAGAVPT SAVPTGRHIQ PFGVGIPIDE YTWNLGTTSR QDIRYIYHS AIDVGARRVN LAIVSRQRAD NVYVLRPTTV ASRPVIGIGL GNDVFLTAHA LASGGPDAAA IVRVTINFFR Q PQMRHLSW ...String: NISDYKVMTW NLQGSSASTE SKWNVNVRQL LSGTAGVDIL MVQEAGAVPT SAVPTGRHIQ PFGVGIPIDE YTWNLGTTSR QDIRYIYHS AIDVGARRVN LAIVSRQRAD NVYVLRPTTV ASRPVIGIGL GNDVFLTAHA LASGGPDAAA IVRVTINFFR Q PQMRHLSW FLAGDFNRSP DRLENDLMTE HLERVVAVLA PTEPTQIGGG ILDYGVIVDR APYSQRVEAL RNPQLASDHY PV AFLARSC LEHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 2.0 second before plunging. | |||||||||
| Details | Freshly prepared size-exclusion-chromatography purified complex of TyTx11 IgG and Typhoid toxin wild-type |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 63000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Initial local fitting was done using Chimera and then Coot was used for rebuilding Fab variable domains into correct sequences. Refinement was performed using Real Space Refine in PHENIX and was iterated with manual building in Coot. | ||||||||
| Output model |  PDB-6vx4: |
 Movie
Movie Controller
Controller



















 Z
Z Y
Y X
X