+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21225 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PilA-N/C from Geobacter sulfurreducens | |||||||||||||||
 Map data Map data | Cryo-EM structure of PilA-N-C from Geobacter sulfurreducens | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Protein transport / Pili / PROTEIN FIBRIL | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpilus assembly / protein secretion by the type II secretion system / type II protein secretion system complex / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Geobacter sulfurreducens (bacteria) Geobacter sulfurreducens (bacteria) | |||||||||||||||
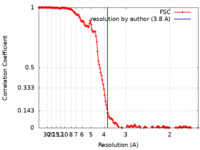
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Gu Y / Srikanth V | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure of Geobacter pili reveals secretory rather than nanowire behaviour. Authors: Yangqi Gu / Vishok Srikanth / Aldo I Salazar-Morales / Ruchi Jain / J Patrick O'Brien / Sophia M Yi / Rajesh Kumar Soni / Fadel A Samatey / Sibel Ebru Yalcin / Nikhil S Malvankar /  Abstract: Extracellular electron transfer by Geobacter species through surface appendages known as microbial nanowires is important in a range of globally important environmental phenomena, as well as for ...Extracellular electron transfer by Geobacter species through surface appendages known as microbial nanowires is important in a range of globally important environmental phenomena, as well as for applications in bio-remediation, bioenergy, biofuels and bioelectronics. Since 2005, these nanowires have been thought to be type 4 pili composed solely of the PilA-N protein. However, previous structural analyses have demonstrated that, during extracellular electron transfer, cells do not produce pili but rather nanowires made up of the cytochromes OmcS and OmcZ. Here we show that Geobacter sulfurreducens binds PilA-N to PilA-C to assemble heterodimeric pili, which remain periplasmic under nanowire-producing conditions that require extracellular electron transfer. Cryo-electron microscopy revealed that C-terminal residues of PilA-N stabilize its copolymerization with PilA-C (to form PilA-N-C) through electrostatic and hydrophobic interactions that position PilA-C along the outer surface of the filament. PilA-N-C filaments lack π-stacking of aromatic side chains and show a conductivity that is 20,000-fold lower than that of OmcZ nanowires. In contrast with surface-displayed type 4 pili, PilA-N-C filaments show structure, function and localization akin to those of type 2 secretion pseudopili. The secretion of OmcS and OmcZ nanowires is lost when pilA-N is deleted and restored when PilA-N-C filaments are reconstituted. The substitution of pilA-N with the type 4 pili of other microorganisms also causes a loss of secretion of OmcZ nanowires. As all major phyla of prokaryotes use systems similar to type 4 pili, this nanowire translocation machinery may have a widespread effect in identifying the evolution and prevalence of diverse electron-transferring microorganisms and in determining nanowire assembly architecture for designing synthetic protein nanowires. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21225.map.gz emd_21225.map.gz | 17.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21225-v30.xml emd-21225-v30.xml emd-21225.xml emd-21225.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21225_fsc.xml emd_21225_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_21225.png emd_21225.png | 175 KB | ||
| Filedesc metadata |  emd-21225.cif.gz emd-21225.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21225 http://ftp.pdbj.org/pub/emdb/structures/EMD-21225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21225 | HTTPS FTP |
-Validation report
| Summary document |  emd_21225_validation.pdf.gz emd_21225_validation.pdf.gz | 415.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21225_full_validation.pdf.gz emd_21225_full_validation.pdf.gz | 414.9 KB | Display | |
| Data in XML |  emd_21225_validation.xml.gz emd_21225_validation.xml.gz | 13.8 KB | Display | |
| Data in CIF |  emd_21225_validation.cif.gz emd_21225_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21225 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21225 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21225 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21225 | HTTPS FTP |
-Related structure data
| Related structure data |  6vk9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10797 (Title: Cryo-EM structure of PilA-N/C from Geobacter sulfurreducens EMPIAR-10797 (Title: Cryo-EM structure of PilA-N/C from Geobacter sulfurreducensData size: 1.5 TB Data #1: Geobacter sulfurreducens PilA-N-C pili - raw unaligned/uncorrected super resolution images [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21225.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21225.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of PilA-N-C from Geobacter sulfurreducens | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Polymerized hetero-dimers of PilA-N and PilA-C
| Entire | Name: Polymerized hetero-dimers of PilA-N and PilA-C |
|---|---|
| Components |
|
-Supramolecule #1: Polymerized hetero-dimers of PilA-N and PilA-C
| Supramolecule | Name: Polymerized hetero-dimers of PilA-N and PilA-C / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Geobacter sulfurreducens (bacteria) Geobacter sulfurreducens (bacteria) |
-Macromolecule #1: Geopilin domain 1 protein
| Macromolecule | Name: Geopilin domain 1 protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Geobacter sulfurreducens (bacteria) Geobacter sulfurreducens (bacteria) |
| Molecular weight | Theoretical: 6.576502 KDa |
| Recombinant expression | Organism:  Geobacter sulfurreducens (bacteria) Geobacter sulfurreducens (bacteria) |
| Sequence | String: FTLIELLIVV AIIGILAAIA IPQFSAYRVK AYNSAASSDL RNLKTALESA FADDQTYPPE S UniProtKB: Geopilin domain 1 protein |
-Macromolecule #2: Geopilin domain 2 protein
| Macromolecule | Name: Geopilin domain 2 protein / type: protein_or_peptide / ID: 2 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Geobacter sulfurreducens (bacteria) Geobacter sulfurreducens (bacteria) |
| Molecular weight | Theoretical: 10.973763 KDa |
| Recombinant expression | Organism:  Geobacter sulfurreducens (bacteria) Geobacter sulfurreducens (bacteria) |
| Sequence | String: AGKIPTTTMG GKDFTFKPST NVSVSYFTTN GATSTAGTVN TDYAVNTKNS SGNRVFTSTN NTSNIWYIEN DAWKGKAVSD SDVTALGTG DVGKSDFSGT EWKSQ UniProtKB: Geopilin domain 2 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.032 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 2-10 / Number grids imaged: 1 / Number real images: 7664 / Average exposure time: 0.2 sec. / Average electron dose: 2.308 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6vk9: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)