[English] 日本語
 Yorodumi
Yorodumi- EMDB-15179: Cryo-EM reconstruction of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80 | ||||||||||||||||||
 Map data Map data | sharpened map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | chromatin remodeler / INO80 / Actin-related protein / DNA binding protein | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcellular bud neck contractile ring / mitotic actomyosin contractile ring contraction / : / RHOA GTPase cycle / vacuole inheritance / ascospore wall assembly / actin cortical patch / mitotic recombination / kinetochore assembly / regulation of TOR signaling ...cellular bud neck contractile ring / mitotic actomyosin contractile ring contraction / : / RHOA GTPase cycle / vacuole inheritance / ascospore wall assembly / actin cortical patch / mitotic recombination / kinetochore assembly / regulation of TOR signaling / Swr1 complex / Ino80 complex / telomere maintenance via recombination / ATP-dependent chromatin remodeler activity / regulation of metabolic process / SWI/SNF complex / cellular response to stress / actin filament bundle / establishment of cell polarity / NuA4 histone acetyltransferase complex / protein secretion / chromosome, centromeric region / subtelomeric heterochromatin formation / actin filament / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / endocytosis / double-strand break repair / actin cytoskeleton / chromatin organization / histone binding / transcription by RNA polymerase II / cytoskeleton / chromosome, telomeric region / chromatin remodeling / DNA repair / mRNA binding / DNA damage response / chromatin binding / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / DNA-templated transcription / chromatin / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | Kunert F / Metzner FJ / Eustermann S / Jung J / Woike S / Schall K / Kostrewa D / Hopfner KP | ||||||||||||||||||
| Funding support | European Union,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structural mechanism of extranucleosomal DNA readout by the INO80 complex. Authors: Franziska Kunert / Felix J Metzner / James Jung / Markus Höpfler / Stephan Woike / Kevin Schall / Dirk Kostrewa / Manuela Moldt / Jia-Xuan Chen / Susanne Bantele / Boris Pfander / Sebastian ...Authors: Franziska Kunert / Felix J Metzner / James Jung / Markus Höpfler / Stephan Woike / Kevin Schall / Dirk Kostrewa / Manuela Moldt / Jia-Xuan Chen / Susanne Bantele / Boris Pfander / Sebastian Eustermann / Karl-Peter Hopfner /  Abstract: The nucleosomal landscape of chromatin depends on the concerted action of chromatin remodelers. The INO80 remodeler specifically places nucleosomes at the boundary of gene regulatory elements, which ...The nucleosomal landscape of chromatin depends on the concerted action of chromatin remodelers. The INO80 remodeler specifically places nucleosomes at the boundary of gene regulatory elements, which is proposed to be the result of an ATP-dependent nucleosome sliding activity that is regulated by extranucleosomal DNA features. Here, we use cryo-electron microscopy and functional assays to reveal how INO80 binds and is regulated by extranucleosomal DNA. Structures of the regulatory A-module bound to DNA clarify the mechanism of linker DNA binding. The A-module is connected to the motor unit via an HSA/post-HSA lever element to chemomechanically couple the motor and linker DNA sensing. Two notable sites of curved DNA recognition by coordinated action of the four actin/actin-related proteins and the motor suggest how sliding by INO80 can be regulated by extranucleosomal DNA features. Last, the structures clarify the recruitment of YY1/Ies4 subunits and reveal deep architectural similarities between the regulatory modules of INO80 and SWI/SNF complexes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15179.map.gz emd_15179.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15179-v30.xml emd-15179-v30.xml emd-15179.xml emd-15179.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15179.png emd_15179.png | 95.6 KB | ||
| Filedesc metadata |  emd-15179.cif.gz emd-15179.cif.gz | 7.4 KB | ||
| Others |  emd_15179_half_map_1.map.gz emd_15179_half_map_1.map.gz emd_15179_half_map_2.map.gz emd_15179_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15179 http://ftp.pdbj.org/pub/emdb/structures/EMD-15179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15179 | HTTPS FTP |
-Related structure data
| Related structure data |  8a5oMC  8a5aC  8a5dC  8a5pC  8a5qC  8atfC  8av6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15179.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15179.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
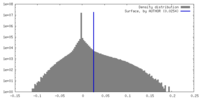
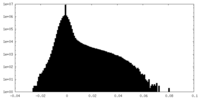
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_15179_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
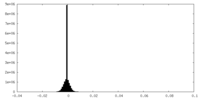
| Density Histograms |
-Half map: half map A
| File | emd_15179_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
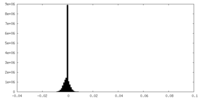
| Density Histograms |
- Sample components
Sample components
-Entire : INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA)
| Entire | Name: INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) |
|---|---|
| Components |
|
-Supramolecule #1: INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA)
| Supramolecule | Name: INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chromatin-remodeling ATPase INO80
| Macromolecule | Name: Chromatin-remodeling ATPase INO80 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.170648 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSLAVLLNKE DKDISDFSKT TAGKSAKKNS RERVADVAPT RVLDKKQAYL SQLNSEFNRI KRRDSIEQLY QDWKFINLQE FELISEWNQ QSKDWQFDNT NDSQDLHFKK LYRDMSMINK EWAEYQSFKN ANLSDIINEK DADEDEEDDE DELEDGEEDM E EDEASTGR ...String: MSLAVLLNKE DKDISDFSKT TAGKSAKKNS RERVADVAPT RVLDKKQAYL SQLNSEFNRI KRRDSIEQLY QDWKFINLQE FELISEWNQ QSKDWQFDNT NDSQDLHFKK LYRDMSMINK EWAEYQSFKN ANLSDIINEK DADEDEEDDE DELEDGEEDM E EDEASTGR HTNGKSMRGN GIQKSRKKDA AAAAAIGKAI KDDQTHADTV VTVNGDENED GNNGEDEDND NDNENNNDND ND NENENDN DSDNDDEEEN GEEDEEEEEI EDLDEEDFAA FEEQDDNDDE DFNPDVEKRR KRSSSSSSST KLSMNSLSLI TSK KINKNI TINSDRPKIV RELIKMCNKN KHQKIKKRRF TNCIVTDYNP IDSKLNIKIT LKQYHVKRLK KLINDAKRER EREE ALKNN VGLDGNDLDN DEDGSESHKR RKLNNNTANG ADDANKRKFN TRHGLPTYGM KMNAKEARAI QRHYDNTYTT IWKDM ARKD STKMSRLVQQ IQSIRSTNFR KTSSLCAREA KKWQSKNFKQ IKDFQTRARR GIREMSNFWK KNEREERDLK KKIEKE AME QAKKEEEEKE SKRQAKKLNF LLTQTELYSH FIGRKDYKDD DDKGTDYKDD DDK UniProtKB: Chromatin-remodeling ATPase INO80 |
-Macromolecule #2: Actin-like protein ARP8
| Macromolecule | Name: Actin-like protein ARP8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 100.322688 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSQEEAESSI IYEEPIDIPL EDDDDEDELE EENSVPLSSQ ADQENAENES DDSVDNVVGS ETPRSVTGLS VDPRDVADEE DEDEEGEDE DEDEDDNDVD NEDENDNDNA NENENELGSS RDKRAPPAVQ TSKRYKKYPK LDPAKAPPGK KVPLHLLEKR R LGRIKAAE ...String: MSQEEAESSI IYEEPIDIPL EDDDDEDELE EENSVPLSSQ ADQENAENES DDSVDNVVGS ETPRSVTGLS VDPRDVADEE DEDEEGEDE DEDEDDNDVD NEDENDNDNA NENENELGSS RDKRAPPAVQ TSKRYKKYPK LDPAKAPPGK KVPLHLLEKR R LGRIKAAE EFAKTLKKIG IEKVETTTLP ATGLFQPLML INQKNYSSDY LKKDDQIFAL RDRKFLRNNN TSQISSTNTP DV IDLKSLP HSEASAAPLN DEIDLNDPTA TIVIHPGSNS IKIGFPKDDH PVVVPNCVAV PKKWLDLENS EHVENVCLQR EQS EEFNNI KSEMEKNFRE RMRYYKRKVP GNAHEQVVSF NENSKPEIIS EKNDPSPIEW IFDDSKLYYG SDALRCVDEK FVIR KPFRG GSFNVKSPYY KSLAELISDV TKLLEHALNS ETLNVKPTKF NQYKVVLVIP DIFKKSHVET FIRVLLTELQ FQAVA IIQE SLATCYGAGI STSTCVVNIG AAETRIACVD EGTVLEHSAI TLDYGGDDIT RLFALFLLQS DFPLQDWKID SKHGWL LAE RLKKNFTTFQ DADVAVQLYN FMNRSPNQPT EKYEFKLFDE VMLAPLALFF PQIFKLIRTS SHKNSSLEFQ LPESRDL FT NELNDWNSLS QFESKEGNLY CDLNDDLKIL NRILDAHNII DQLQDKPENY GNTLKENFAP LEKAIVQSIA NASITADV T RMNSFYSNIL IVGGSSKIPA LDFILTDRIN IWRPSLLSSA SFPQFYKKLT KEIKDLEGHY VNAPDKTEDE NKQILQAQI KEKIVEELEE QHQNIEHQNG NEHIFPVSII PPPRDMNPAL IIWKGASVLA QIKLVEELFI TNSDWDVHGS RILQYKCIFT Y UniProtKB: Actin-like protein ARP8 |
-Macromolecule #3: Actin
| Macromolecule | Name: Actin / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.735547 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDSEVAALVI DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGIMVGMGQK DSYVGDEAQS KRGILTLRYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP MNPKSNREKM TQIMFETFNV PAFYVSIQAV LSLYSSGRTT GIVLDSGDGV T HVVPIYAG ...String: MDSEVAALVI DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGIMVGMGQK DSYVGDEAQS KRGILTLRYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP MNPKSNREKM TQIMFETFNV PAFYVSIQAV LSLYSSGRTT GIVLDSGDGV T HVVPIYAG FSLPHAILRI DLAGRDLTDY LMKILSERGY SFSTTAEREI VRDIKEKLCY VALDFEQEMQ TAAQSSSIEK SY ELPDGQV ITIGNERFRA PEALFHPSVL GLESAGIDQT TYNSIMKCDV DVRKELYGNI VMSGGTTMFP GIAERMQKEI TAL APSSMK VKIIAPPERK YSVWIGGSIL ASLTTFQQMW ISKQEYDESG PSIVHHKCF UniProtKB: Actin |
-Macromolecule #4: Actin-related protein 4
| Macromolecule | Name: Actin-related protein 4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.894684 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSNAALQVYG GDEVSAVVID PGSYTTNIGY SGSDFPQSIL PSVYGKYTAD EGNKKIFSEQ SIGIPRKDYE LKPIIENGLV IDWDTAQEQ WQWALQNELY LNSNSGIPAL LTEPVWNSTE NRKKSLEVLL EGMQFEACYL APTSTCVSFA AGRPNCLVVD I GHDTCSVS ...String: MSNAALQVYG GDEVSAVVID PGSYTTNIGY SGSDFPQSIL PSVYGKYTAD EGNKKIFSEQ SIGIPRKDYE LKPIIENGLV IDWDTAQEQ WQWALQNELY LNSNSGIPAL LTEPVWNSTE NRKKSLEVLL EGMQFEACYL APTSTCVSFA AGRPNCLVVD I GHDTCSVS PIVDGMTLSK STRRNFIAGK FINHLIKKAL EPKEIIPLFA IKQRKPEFIK KTFDYEVDKS LYDYANNRGF FQ ECKETLC HICPTKTLEE TKTELSSTAK RSIESPWNEE IVFDNETRYG FAEELFLPKE DDIPANWPRS NSGVVKTWRN DYV PLKRTK PSGVNKSDKK VTPTEEKEQE AVSKSTSPAA NSADTPNETG KRPLEEEKPP KENNELIGLA DLVYSSIMSS DVDL RATLA HNVVLTGGTS SIPGLSDRLM TELNKILPSL KFRILTTGHT IERQYQSWLG GSILTSLGTF HQLWVGKKEY EEVGV ERLL NDRFR UniProtKB: Actin-related protein 4 |
-Macromolecule #5: Ino eighty subunit 4
| Macromolecule | Name: Ino eighty subunit 4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.111197 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSQESSVLSE SQEQLANNPK IEDTSPPSAN SRDNSKPVLP WDYKNKAIEI KSFSGYKVNF TGWIRRDVRE ERQRGSEFTA SDVKGSDDK ATRKKEPADE DPEVKQLEKE GEDGLDS UniProtKB: Ino eighty subunit 4 |
-Macromolecule #6: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 6 / Number of copies: 3 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 47.37 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 250000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)