[English] 日本語
 Yorodumi
Yorodumi- EMDB-14517: Chimaera of AP2 with FCHO2 linker domain as a fusion on Cmu2 subunit -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chimaera of AP2 with FCHO2 linker domain as a fusion on Cmu2 subunit | |||||||||
 Map data Map data | Local resolution filtered overall map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adaptor complex / AP2 / trafficking / clathrin / CCP / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationLDL clearance / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / Gap junction degradation / Formation of annular gap junctions / WNT5A-dependent internalization of FZD4 / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / Nef Mediated CD8 Down-regulation / Trafficking of GluR2-containing AMPA receptors / LDL clearance / Retrograde neurotrophin signalling ...LDL clearance / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / Gap junction degradation / Formation of annular gap junctions / WNT5A-dependent internalization of FZD4 / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / Nef Mediated CD8 Down-regulation / Trafficking of GluR2-containing AMPA receptors / LDL clearance / Retrograde neurotrophin signalling / Retrograde neurotrophin signalling / VLDLR internalisation and degradation / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / WNT5A-dependent internalization of FZD4 / Trafficking of GluR2-containing AMPA receptors / clathrin adaptor complex / VLDLR internalisation and degradation / WNT5A-dependent internalization of FZD4 / Recycling pathway of L1 / extrinsic component of presynaptic endocytic zone membrane / MHC class II antigen presentation / regulation of vesicle size / postsynaptic endocytic zone / AP-2 adaptor complex / postsynaptic neurotransmitter receptor internalization / Cargo recognition for clathrin-mediated endocytosis / Recycling pathway of L1 / Retrograde neurotrophin signalling / clathrin-coated endocytic vesicle / membrane coat / Clathrin-mediated endocytosis / clathrin coat assembly / positive regulation of synaptic vesicle endocytosis / Cargo recognition for clathrin-mediated endocytosis / clathrin-cargo adaptor activity / Clathrin-mediated endocytosis / LDL clearance / vesicle budding from membrane / clathrin-dependent endocytosis / MHC class II antigen presentation / signal sequence binding / coronary vasculature development / Nef Mediated CD4 Down-regulation / positive regulation of protein localization to membrane / endolysosome membrane / neurotransmitter secretion / Neutrophil degranulation / aorta development / ventricular septum development / low-density lipoprotein particle receptor binding / clathrin binding / Trafficking of GluR2-containing AMPA receptors / Recycling pathway of L1 / positive regulation of receptor internalization / EPH-ephrin mediated repulsion of cells / positive regulation of endocytosis / synaptic vesicle endocytosis / negative regulation of protein localization to plasma membrane / vesicle-mediated transport / clathrin-coated pit / phosphatidylinositol binding / MHC class II antigen presentation / protein serine/threonine kinase binding / VLDLR internalisation and degradation / intracellular protein transport / kidney development / clathrin-coated endocytic vesicle membrane / receptor internalization / cytoplasmic side of plasma membrane / kinase binding / terminal bouton / disordered domain specific binding / synaptic vesicle / endocytic vesicle membrane / Cargo recognition for clathrin-mediated endocytosis / presynapse / Clathrin-mediated endocytosis / protein-containing complex assembly / cytoplasmic vesicle / Potential therapeutics for SARS / transmembrane transporter binding / postsynapse / protein domain specific binding / synapse / lipid binding / protein kinase binding / protein-containing complex binding / glutamatergic synapse / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |    Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
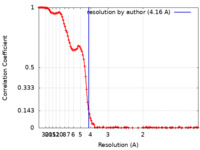
| Method | single particle reconstruction / cryo EM / Resolution: 4.16 Å | |||||||||
 Authors Authors | Kane Dickson V / Qu K / Owen DJ / Briggs JA / Zaccai NR | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P-dependent switch. Authors: Nathan R Zaccai / Zuzana Kadlecova / Veronica Kane Dickson / Kseniya Korobchevskaya / Jan Kamenicky / Oleksiy Kovtun / Perunthottathu K Umasankar / Antoni G Wrobel / Jonathan G G Kaufman / ...Authors: Nathan R Zaccai / Zuzana Kadlecova / Veronica Kane Dickson / Kseniya Korobchevskaya / Jan Kamenicky / Oleksiy Kovtun / Perunthottathu K Umasankar / Antoni G Wrobel / Jonathan G G Kaufman / Sally R Gray / Kun Qu / Philip R Evans / Marco Fritzsche / Filip Sroubek / Stefan Höning / John A G Briggs / Bernard T Kelly / David J Owen / Linton M Traub /      Abstract: Clathrin-mediated endocytosis (CME) is the main mechanism by which mammalian cells control their cell surface proteome. Proper operation of the pivotal CME cargo adaptor AP2 requires membrane- ...Clathrin-mediated endocytosis (CME) is the main mechanism by which mammalian cells control their cell surface proteome. Proper operation of the pivotal CME cargo adaptor AP2 requires membrane-localized Fer/Cip4 homology domain-only proteins (FCHO). Here, live-cell enhanced total internal reflection fluorescence-structured illumination microscopy shows that FCHO marks sites of clathrin-coated pit (CCP) initiation, which mature into uniform-sized CCPs comprising a central patch of AP2 and clathrin corralled by an FCHO/Epidermal growth factor potential receptor substrate number 15 (Eps15) ring. We dissect the network of interactions between the FCHO interdomain linker and AP2, which concentrates, orients, tethers, and partially destabilizes closed AP2 at the plasma membrane. AP2's subsequent membrane deposition drives its opening, which triggers FCHO displacement through steric competition with phosphatidylinositol 4,5-bisphosphate, clathrin, cargo, and CME accessory factors. FCHO can now relocate toward a CCP's outer edge to engage and activate further AP2s to drive CCP growth/maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14517.map.gz emd_14517.map.gz | 11 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14517-v30.xml emd-14517-v30.xml emd-14517.xml emd-14517.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14517_fsc.xml emd_14517_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14517.png emd_14517.png | 125.6 KB | ||
| Filedesc metadata |  emd-14517.cif.gz emd-14517.cif.gz | 7.6 KB | ||
| Others |  emd_14517_half_map_1.map.gz emd_14517_half_map_1.map.gz emd_14517_half_map_2.map.gz emd_14517_half_map_2.map.gz | 139 MB 139 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14517 http://ftp.pdbj.org/pub/emdb/structures/EMD-14517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14517 | HTTPS FTP |
-Related structure data
| Related structure data |  7z5cMC  7ofpC  7og1C  7ohiC  7ohoC  7ohzC  7oi5C  7oiqC  7oitC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14517.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14517.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered overall map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.652 Å | ||||||||||||||||||||||||||||||||||||
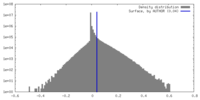
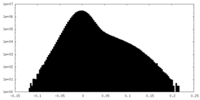
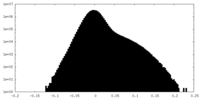
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_14517_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
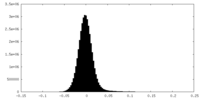
| Density Histograms |
-Half map: Half map B
| File | emd_14517_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
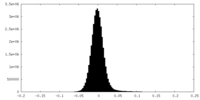
| Density Histograms |
- Sample components
Sample components
-Entire : AP2 core complex
| Entire | Name: AP2 core complex |
|---|---|
| Components |
|
-Supramolecule #1: AP2 core complex
| Supramolecule | Name: AP2 core complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Ternary complex of AP2 core expressed as part of a chimaera with FCHO2 linker (not modelled) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 203 KDa |
-Macromolecule #1: AP-2 complex subunit alpha-2
| Macromolecule | Name: AP-2 complex subunit alpha-2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.656297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPAVSKGEGM RGLAVFISDI RNCKSKEAEI KRINKELANI RSKFKGDKAL DGYSKKKYVC KLLFIFLLGH DIDFGHMEAV NLLSSNRYT EKQIGYLFIS VLVNSNSELI RLINNAIKND LASRNPTFMG LALHCIANVG SREMAEAFAG EIPKILVAGD T MDSVKQSA ...String: MPAVSKGEGM RGLAVFISDI RNCKSKEAEI KRINKELANI RSKFKGDKAL DGYSKKKYVC KLLFIFLLGH DIDFGHMEAV NLLSSNRYT EKQIGYLFIS VLVNSNSELI RLINNAIKND LASRNPTFMG LALHCIANVG SREMAEAFAG EIPKILVAGD T MDSVKQSA ALCLLRLYRT SPDLVPMGDW TSRVVHLLND QHLGVVTAAT SLITTLAQKN PEEFKTSVSL AVSRLSRIVT SA STDLQDY TYYFVPAPWL SVKLLRLLQC YPPPEDPAVR GRLTECLETI LNKAQEPPKS KKVQHSNAKN AVLFEAISLI IHH DSEPNL LVRACNQLGQ FLQHRETNLR YLALESMCTL ASSEFSHEAV KTHIETVINA LKTERDVSVR QRAVDLLYAM CDRS NAQQI VAEMLSYLET ADYSIREEIV LKVAILAEKY AVDYTWYVDT ILNLIRIAGD YVSEEVWYRV IQIVINRDDV QGYAA KTVF EALQAPACHE NLVKVGGYIL GEFGNLIAGD PRSSPLIQFN LLHSKFHLCS VPTRALLLST YIKFVNLFPE VKATIQ DVL RSDSQLKNAD VELQQRAVEY LRLSTVASTD ILATVLEEMP PFPERESSIL AKLKKKKG UniProtKB: AP-2 complex subunit alpha-2 |
-Macromolecule #2: AP-2 complex subunit beta
| Macromolecule | Name: AP-2 complex subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.953195 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDSKYFTTN KKGEIFELKA ELNNEKKEKR KEAVKKVIAA MTVGKDVSSL FPDVVNCMQT DNLELKKLVY LYLMNYAKSQ PDMAIMAVN SFVKDCEDPN PLIRALAVRT MGCIRVDKIT EYLCEPLRKC LKDEDPYVRK TAAVCVAKLH DINAQMVEDQ G FLDSLRDL ...String: MTDSKYFTTN KKGEIFELKA ELNNEKKEKR KEAVKKVIAA MTVGKDVSSL FPDVVNCMQT DNLELKKLVY LYLMNYAKSQ PDMAIMAVN SFVKDCEDPN PLIRALAVRT MGCIRVDKIT EYLCEPLRKC LKDEDPYVRK TAAVCVAKLH DINAQMVEDQ G FLDSLRDL IADSNPMVVA NAVAALSEIS ESHPNSNLLD LNPQNINKLL TALNECTEWG QIFILDCLSN YNPKDDREAQ SI CERVTPR LSHANSAVVL SAVKVLMKFL ELLPKDSDYY NMLLKKLAPP LVTLLSGEPE VQYVALRNIN LIVQKRPEIL KQE IKVFFV KYNDPIYVKL EKLDIMIRLA SQANIAQVLA ELKEYATEVD VDFVRKAVRA IGRCAIKVEQ SAERCVSTLL DLIQ TKVNY VVQEAIVVIR DIFRKYPNKY ESIIATLCEN LDSLDEPDAR AAMIWIVGEY AERIDNADEL LESFLEGFHD ESTQV QLTL LTAIVKLFLK KPSETQELVQ QVLSLATQDS DNPDLRDRGY IYWRLLSTDP VTAKEVVLSE KPLISEETDL IEPTLL DEL ICHIGSLASV YHKPPNAFVE GSHGIHRK UniProtKB: AP-2 complex subunit beta |
-Macromolecule #3: AP-2 complex subunit mu
| Macromolecule | Name: AP-2 complex subunit mu / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.726641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIGGLFIYNH KGEVLISRVY RDDIGRNAVD AFRVNVIHAR QQVRSPVTNI ARTSFFHVKR SNIWLAAVTK QNVNAAMVFE FLYKMCDVM AAYFGKISEE NIKNNFVLIY ELLDEILDFG YPQNSETGAL KTFITQQGIK SQHQTKEEQS QITSQVTGQI G WRREGIKY ...String: MIGGLFIYNH KGEVLISRVY RDDIGRNAVD AFRVNVIHAR QQVRSPVTNI ARTSFFHVKR SNIWLAAVTK QNVNAAMVFE FLYKMCDVM AAYFGKISEE NIKNNFVLIY ELLDEILDFG YPQNSETGAL KTFITQQGIK SQHQTKEEQS QITSQVTGQI G WRREGIKY RRNELFLDVL ESVNLLMSPQ GQVLSAHVSG RVVMKSYLSG MPECKFGMND KIVIEKQGKG TADETSKSGK QS IAIDDCT FHQCVRLSKF DSERSISFIP PDGEFELMRY RTTKDIILPF RVIPLVREVG RTKLEVKVVI KSNFKPSLLA QKI EVRIPT PLNTSGVQVI CMKGKAKYKA SENAIVWKIK RMAGMKESQI SAEIELLPTN DKKKWARPPI SMNFEVPFAP SGLK VRYLK VFEPKLNYSD HDVIKWVRYI GRSGIYETRC UniProtKB: AP-2 complex subunit mu |
-Macromolecule #4: AP-2 complex subunit sigma
| Macromolecule | Name: AP-2 complex subunit sigma / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.038688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIRFILIQNR AGKTRLAKWY MQFDDDEKQK LIEEVHAVVT VRDAKHTNFV EFRNFKIIYR RYAGLYFCIC VDVNDNNLAY LEAIHNFVE VLNEYFHNVC ELDLVFNFYK VYTVVDEMFL AGEIRETSQT KVLKQLLMLQ SLE UniProtKB: AP-2 complex subunit sigma |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 50% HKT buffer (10mM Hepes, 10mM Tris 120mM potassium acetate pH 7.2) and 50% Core buffer (10mM Tris, 250mM NaCl, pH 8) |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)