[English] 日本語
 Yorodumi
Yorodumi- EMDB-1412: Interaction of decay-accelerating factor with coxsackievirus B3. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1412 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Interaction of decay-accelerating factor with coxsackievirus B3. | |||||||||
 Map data Map data | Surface rendered cvb3 complexed with DAF. Calculated map was used to scaled complexmap to 2.92 for fitting of DAF into the cryoEM density. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of lipopolysaccharide-mediated signaling pathway / negative regulation of complement activation / regulation of complement-dependent cytotoxicity / regulation of complement activation / respiratory burst / positive regulation of CD4-positive, alpha-beta T cell activation / positive regulation of CD4-positive, alpha-beta T cell proliferation / Class B/2 (Secretin family receptors) / ficolin-1-rich granule membrane / complement activation, classical pathway ...regulation of lipopolysaccharide-mediated signaling pathway / negative regulation of complement activation / regulation of complement-dependent cytotoxicity / regulation of complement activation / respiratory burst / positive regulation of CD4-positive, alpha-beta T cell activation / positive regulation of CD4-positive, alpha-beta T cell proliferation / Class B/2 (Secretin family receptors) / ficolin-1-rich granule membrane / complement activation, classical pathway / transport vesicle / side of membrane / COPI-mediated anterograde transport / endoplasmic reticulum-Golgi intermediate compartment membrane / secretory granule membrane / Regulation of Complement cascade / positive regulation of T cell cytokine production / virus receptor activity / positive regulation of cytosolic calcium ion concentration / membrane raft / Golgi membrane / innate immune response / lipid binding / Neutrophil degranulation / cell surface / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / coxsackievirus B3 passaged through rhabdomysarcoma cells rhabdomyosarcoma cells Homo sapiens (human) / coxsackievirus B3 passaged through rhabdomysarcoma cells rhabdomyosarcoma cells | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 14.0 Å | |||||||||
 Authors Authors | Hafenstein S | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2007 Journal: J Virol / Year: 2007Title: Interaction of decay-accelerating factor with coxsackievirus B3. Authors: Susan Hafenstein / Valorie D Bowman / Paul R Chipman / Carol M Bator Kelly / Feng Lin / M Edward Medof / Michael G Rossmann /  Abstract: Many entero-, parecho-, and rhinoviruses use immunoglobulin (Ig)-like receptors that bind into the viral canyon and are required to initiate viral uncoating during infection. However, some of these ...Many entero-, parecho-, and rhinoviruses use immunoglobulin (Ig)-like receptors that bind into the viral canyon and are required to initiate viral uncoating during infection. However, some of these viruses use an alternative or additional receptor that binds outside the canyon. Both the coxsackievirus-adenovirus receptor (CAR), an Ig-like molecule that binds into the viral canyon, and decay-accelerating factor (DAF) have been identified as cellular receptors for coxsackievirus B3 (CVB3). A cryoelectron microscopy reconstruction of a variant of CVB3 complexed with DAF shows full occupancy of the DAF receptor in each of 60 binding sites. The DAF molecule bridges the canyon, blocking the CAR binding site and causing the two receptors to compete with one another. The binding site of DAF on CVB3 differs from the binding site of DAF on the surface of echoviruses, suggesting independent evolutionary processes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1412.map.gz emd_1412.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1412-v30.xml emd-1412-v30.xml emd-1412.xml emd-1412.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1412.gif 1412.gif | 129.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1412 http://ftp.pdbj.org/pub/emdb/structures/EMD-1412 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1412 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1412 | HTTPS FTP |
-Validation report
| Summary document |  emd_1412_validation.pdf.gz emd_1412_validation.pdf.gz | 350.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1412_full_validation.pdf.gz emd_1412_full_validation.pdf.gz | 350.5 KB | Display | |
| Data in XML |  emd_1412_validation.xml.gz emd_1412_validation.xml.gz | 5.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1412 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1412 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1412 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1412 | HTTPS FTP |
-Related structure data
| Related structure data |  2qzdMC  2qzfMC  2qzhMC  1411C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1412.map.gz / Format: CCP4 / Size: 22.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1412.map.gz / Format: CCP4 / Size: 22.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Surface rendered cvb3 complexed with DAF. Calculated map was used to scaled complexmap to 2.92 for fitting of DAF into the cryoEM density. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.92 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
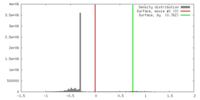
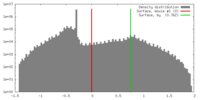
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : coxsackievirus B3, RD strain, complexed with decay-accelerating factor
| Entire | Name: coxsackievirus B3, RD strain, complexed with decay-accelerating factor |
|---|---|
| Components |
|
-Supramolecule #1000: coxsackievirus B3, RD strain, complexed with decay-accelerating factor
| Supramolecule | Name: coxsackievirus B3, RD strain, complexed with decay-accelerating factor type: sample / ID: 1000 Oligomeric state: one DAF binds each binding site, one per each protomer Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 7.0 MDa / Theoretical: 7.0 MDa |
-Supramolecule #1: coxsackievirus B3 passaged through rhabdomysarcoma cells rhabdomy...
| Supramolecule | Name: coxsackievirus B3 passaged through rhabdomysarcoma cells rhabdomyosarcoma cells type: virus / ID: 1 / Name.synonym: CVB3-RD Details: virus are saturated with one DAF bound at each possible site Sci species name: coxsackievirus B3 passaged through rhabdomysarcoma cells rhabdomyosarcoma cells Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No / Syn species name: CVB3-RD |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Molecular weight | Experimental: 7.0 MDa / Theoretical: 7.0 MDa |
| Virus shell | Shell ID: 1 / Name: VP1 / Diameter: 300 Å / T number (triangulation number): 1 |
| Virus shell | Shell ID: 2 / Name: VP2 / Diameter: 300 Å / T number (triangulation number): 1 |
| Virus shell | Shell ID: 3 / Name: VP3 / Diameter: 300 Å / T number (triangulation number): 1 |
-Macromolecule #1: decay-accelerating factor
| Macromolecule | Name: decay-accelerating factor / type: protein_or_peptide / ID: 1 / Name.synonym: DAF Details: each molecule of DAF 70kD; DAF has His6 tag at c-terminus Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Experimental: 1.0 MDa / Theoretical: 1.0 MDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 6 / Details: 50mM MES |
| Grid | Details: quantifoils |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 120 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: plunger / Method: blot before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Temperature | Min: 83 K / Max: 83 K / Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: lens astigmatism was corrected at 98,000 times mag |
| Date | Aug 6, 2004 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 44 / Average electron dose: 24 e/Å2 / Details: scanned at 7 microns and bin averaged to 14 / Od range: 1 / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Calibrated magnification: 47000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder: side mounted nitrogen cooled / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | Virion were incubated with DAF at ratio of 4 DAF molecules per every potential binding site. |
|---|---|
| CTF correction | Details: robEM |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 14.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMPFT, EM3DR / Number images used: 2269 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera











 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)