+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13168 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SbmA-FabS11-1 in lipid nanodisc | ||||||||||||||||||||||||
 Map data Map data | cryoSPARC post-processed map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsecondary active transmembrane transporter activity / microcin transmembrane transporter activity / microcin B17 transport / microcin transport / peptide transport / peptide transmembrane transporter activity / protein transport / response to antibiotic / protein homodimerization activity / ATP binding / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
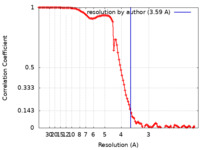
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | ||||||||||||||||||||||||
 Authors Authors | Ghilarov D / Beis K | ||||||||||||||||||||||||
| Funding support |  Poland, Poland,  United Kingdom, United Kingdom,  Japan, Japan,  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Molecular mechanism of SbmA, a promiscuous transporter exploited by antimicrobial peptides. Authors: Dmitry Ghilarov / Satomi Inaba-Inoue / Piotr Stepien / Feng Qu / Elizabeth Michalczyk / Zuzanna Pakosz / Norimichi Nomura / Satoshi Ogasawara / Graham Charles Walker / Sylvie Rebuffat / So ...Authors: Dmitry Ghilarov / Satomi Inaba-Inoue / Piotr Stepien / Feng Qu / Elizabeth Michalczyk / Zuzanna Pakosz / Norimichi Nomura / Satoshi Ogasawara / Graham Charles Walker / Sylvie Rebuffat / So Iwata / Jonathan Gardiner Heddle / Konstantinos Beis /      Abstract: Antibiotic metabolites and antimicrobial peptides mediate competition between bacterial species. Many of them hijack inner and outer membrane proteins to enter cells. Sensitivity of enteric bacteria ...Antibiotic metabolites and antimicrobial peptides mediate competition between bacterial species. Many of them hijack inner and outer membrane proteins to enter cells. Sensitivity of enteric bacteria to multiple peptide antibiotics is controlled by the single inner membrane protein SbmA. To establish the molecular mechanism of peptide transport by SbmA and related BacA, we determined their cryo–electron microscopy structures at 3.2 and 6 Å local resolution, respectively. The structures show a previously unknown fold, defining a new class of secondary transporters named SbmA-like peptide transporters. The core domain includes conserved glutamates, which provide a pathway for proton translocation, powering transport. The structures show an outward-open conformation with a large cavity that can accommodate diverse substrates. We propose a molecular mechanism for antibacterial peptide uptake paving the way for creation of narrow-targeted therapeutics. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13168.map.gz emd_13168.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13168-v30.xml emd-13168-v30.xml emd-13168.xml emd-13168.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13168_fsc.xml emd_13168_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13168.png emd_13168.png | 57 KB | ||
| Others |  emd_13168_additional_1.map.gz emd_13168_additional_1.map.gz emd_13168_half_map_1.map.gz emd_13168_half_map_1.map.gz emd_13168_half_map_2.map.gz emd_13168_half_map_2.map.gz | 105 MB 115.8 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13168 http://ftp.pdbj.org/pub/emdb/structures/EMD-13168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13168 | HTTPS FTP |
-Related structure data
| Related structure data |  7p34MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10777 (Title: Proton-powered peptide transporter SbmA in lipid nanodisc complexed with Fab S11-1 (SbmA-FabS11-1-MccB17) EMPIAR-10777 (Title: Proton-powered peptide transporter SbmA in lipid nanodisc complexed with Fab S11-1 (SbmA-FabS11-1-MccB17)Data size: 2.5 TB Data #1: SbmA-FabS11-1 complex in lipid nanodisc [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13168.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13168.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02125 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: DeepEMhancer post-processed map
| File | emd_13168_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer post-processed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map
| File | emd_13168_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map
| File | emd_13168_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of proton-driven peptide transporter SbmA with Fab S11-1
| Entire | Name: Complex of proton-driven peptide transporter SbmA with Fab S11-1 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of proton-driven peptide transporter SbmA with Fab S11-1
| Supramolecule | Name: Complex of proton-driven peptide transporter SbmA with Fab S11-1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Proton-driven peptide transporter SbmA
| Macromolecule | Name: Proton-driven peptide transporter SbmA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MFKSFFPKPG TFFLSAFVWA LIAVIFWQAG GGDWVARITG ASGQIPISAA RFWSLDFLIF YAYYIVCVG LFALFWFIYS PHRWQYWSIL GTALIIFVTW FLVEVGVAVN AWYAPFYDLI Q TALSSPHK VTIEQFYREV GVFLGIALIA VVISVLNNFF VSHYVFRWRT ...String: MFKSFFPKPG TFFLSAFVWA LIAVIFWQAG GGDWVARITG ASGQIPISAA RFWSLDFLIF YAYYIVCVG LFALFWFIYS PHRWQYWSIL GTALIIFVTW FLVEVGVAVN AWYAPFYDLI Q TALSSPHK VTIEQFYREV GVFLGIALIA VVISVLNNFF VSHYVFRWRT AMNEYYMANW QQ LRHIEGA AQRVQEDTMR FASTLENMGV SFINAIMTLI AFLPVLVTLS AHVPELPIIG HIP YGLVIA AIVWSLMGTG LLAVVGIKLP GLEFKNQRVE AAYRKELVYG EDDATRATPP TVRE LFSAV RKNYFRLYFH YMYFNIARIL YLQVDNVFGL FLLFPSIVAG TITLGLMTQI TNVFG QVRG AFQYLINSWT TLVELMSIYK RLRSFEHELD GDKIQEVTHT LS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial model generated by Buccaneer and refined by PHENIX |
|---|---|
| Output model |  PDB-7p34: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)