Yorodumi
Yorodumi+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11807 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | |||||||||||||||
 Map data Map data | Local resolution filtered | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationrRNA cytidine N-acetyltransferase activity / Butyrate Response Factor 1 (BRF1) binds and destabilizes mRNA / Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA / TRAMP complex / nuclear polyadenylation-dependent mRNA catabolic process / mRNA decay by 3' to 5' exoribonuclease / Noc4p-Nop14p complex / nuclear mRNA surveillance of mRNA 3'-end processing / rRNA acetylation involved in maturation of SSU-rRNA / box H/ACA snoRNA binding ...rRNA cytidine N-acetyltransferase activity / Butyrate Response Factor 1 (BRF1) binds and destabilizes mRNA / Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA / TRAMP complex / nuclear polyadenylation-dependent mRNA catabolic process / mRNA decay by 3' to 5' exoribonuclease / Noc4p-Nop14p complex / nuclear mRNA surveillance of mRNA 3'-end processing / rRNA acetylation involved in maturation of SSU-rRNA / box H/ACA snoRNA binding / regulatory ncRNA 3'-end processing / tRNA N-acetyltransferase activity / regulation of ribosomal protein gene transcription by RNA polymerase II / U1 snRNA 3'-end processing / t-UTP complex / rRNA small subunit pseudouridine methyltransferase Nep1 / tRNA acetylation / U5 snRNA 3'-end processing / RNA fragment catabolic process / CURI complex / UTP-C complex / nuclear polyadenylation-dependent CUT catabolic process / rRNA 2'-O-methylation / Pwp2p-containing subcomplex of 90S preribosome / TRAMP-dependent tRNA surveillance pathway / CUT catabolic process / cytoplasmic exosome (RNase complex) / U4 snRNA 3'-end processing / nuclear polyadenylation-dependent rRNA catabolic process / poly(A)-dependent snoRNA 3'-end processing / histone H2AQ104 methyltransferase activity / exosome (RNase complex) / endonucleolytic cleavage in ITS1 upstream of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / nuclear microtubule / Mpp10 complex / snoRNA guided rRNA 2'-O-methylation / nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay / rRNA (pseudouridine) methyltransferase activity / rRNA modification / nuclear exosome (RNase complex) / nuclear-transcribed mRNA catabolic process, non-stop decay / exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of rRNA processing / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D sno(s)RNA binding / septum digestion after cytokinesis / tRNA re-export from nucleus / snRNA binding / box C/D sno(s)RNA 3'-end processing / regulation of transcription by RNA polymerase I / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA catabolic process / rRNA methyltransferase activity / 3'-5' RNA helicase activity / nuclear mRNA surveillance / rDNA heterochromatin / positive regulation of rRNA processing / single-stranded telomeric DNA binding / box C/D methylation guide snoRNP complex / tRNA export from nucleus / U4/U6 snRNP / rRNA base methylation / mRNA 3'-UTR AU-rich region binding / 90S preribosome assembly / Negative regulators of DDX58/IFIH1 signaling / poly(A) binding / rRNA primary transcript binding / SUMOylation of RNA binding proteins / sno(s)RNA-containing ribonucleoprotein complex / protein localization to nucleolus / Hydrolases; Acting on ester bonds; Endoribonucleases producing 5'-phosphomonoesters / U4 snRNA binding / mTORC1-mediated signalling / O-methyltransferase activity / Protein hydroxylation / rRNA methylation / RNA catabolic process / poly(U) RNA binding / U3 snoRNA binding / mRNA modification / U4 snRNP / maturation of 5.8S rRNA / nonfunctional rRNA decay / positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / poly(A)+ mRNA export from nucleus / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / preribosome, small subunit precursor / Ribosomal scanning and start codon recognition / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / snoRNA binding / precatalytic spliceosome / establishment of cell polarity / positive regulation of transcription by RNA polymerase I / rRNA metabolic process / Major pathway of rRNA processing in the nucleolus and cytosol / nucleolar large rRNA transcription by RNA polymerase I / spliceosomal complex assembly / SRP-dependent cotranslational protein targeting to membrane Similarity search - Function Regulator of rDNA transcription 14 / : / Regular of rDNA transcription protein 14 / Exosome complex component CSL4, N-terminal domain / Exosome complex exonuclease Rrp40, N-terminal / Exosome complex exonuclease Rrp40 N-terminal domain / Apoptosis-antagonizing transcription factor, C-terminal / AATF leucine zipper-containing domain / Protein AATF/Bfr2 / : ...Regulator of rDNA transcription 14 / : / Regular of rDNA transcription protein 14 / Exosome complex component CSL4, N-terminal domain / Exosome complex exonuclease Rrp40, N-terminal / Exosome complex exonuclease Rrp40 N-terminal domain / Apoptosis-antagonizing transcription factor, C-terminal / AATF leucine zipper-containing domain / Protein AATF/Bfr2 / : / Apoptosis-antagonizing transcription factor, C-terminal / Apoptosis antagonizing transcription factor / Exosome complex exonuclease RRP44, S1 domain / S1 domain / NUC153 / Nucleolar protein 10/Enp2 / NUC153 domain / PIN domain / U3 small nucleolar RNA-associated protein 8 / : / Utp8, N-terminal beta propeller / Utp8, C-terminal / Exosome complex component RRP45 / Rrp40, S1 domain / : / Exosome complex component RRP40, S1 domain / rRNA biogenesis protein Rrp5 / : / : / Exosome complex component CSL4, C-terminal / Exosome complex component, N-terminal domain / Exosome complex component Csl4 / Exosome component EXOSC1/CSL4 / Exosome complex exonuclease RRP4 N-terminal region / RRP4, S1 domain / Rrp44-like cold shock domain / Rrp44-like cold shock domain / Ribosomal RNA assembly KRR1 / : / : / : / : / KH domain / KRR1 small subunit processome component, second KH domain / Exosome complex RNA-binding protein 1/RRP40/RRP4 / Dis3-like cold-shock domain 2 / Dis3-like cold-shock domain 2 (CSD2) / U3 small nucleolar RNA-associated protein 20, N-terminal / U3 snoRNA associated / Nucleolar complex protein 4 / U3 small nucleolar RNA-associated protein 20, C-terminal / : / U3 small nucleolar RNA-associated protein 20, N-terminal / U3 snoRNA associated / U3 small nucleolar RNA-associated protein 20, C-terminal / Possible tRNA binding domain / RNA cytidine acetyltransferase NAT10 / Possible tRNA binding domain / Helicase domain / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / TmcA/NAT10/Kre33 / Helicase / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / GNAT acetyltransferase 2 / Nucleolar protein 14 / Nop14-like family / U3 small nucleolar RNA-associated protein 6 / : / U3 small nucleolar RNA-associated protein 6 / NOL6/Upt22 / Small-subunit processome, Utp11 / Sof1-like protein / BING4, C-terminal domain / Fcf2 pre-rRNA processing, C-terminal / Nrap protein domain 1 / Nrap protein, domain 2 / Nrap protein, domain 3 / Nrap protein, domain 4 / Nrap protein, domain 5 / Nrap protein, domain 6 / rRNA-processing protein Fcf1, PIN domain / Fcf2/DNTTIP2 / WD repeat-containing protein WDR46/Utp7 / : / : / Nrap protein domain 1 / Utp11 protein / Sof1-like domain / BING4CT (NUC141) domain / Fcf2 pre-rRNA processing / Nrap protein PAP/OAS-like domain / Nrap protein domain 3 / Nrap protein nucleotidyltransferase domain 4 / Nrap protein PAP/OAS1-like domain 5 / Nrap protein domain 6 / BING4CT (NUC141) domain / rRNA-processing protein Fcf1/Utp23 / Ribonuclease II/R, conserved site / Ribosomal RNA-processing protein 7, C-terminal domain / rRNA-processing arch domain Similarity search - Domain/homology Small ribosomal subunit protein uS4A / Small ribosomal subunit protein uS15 / Small ribosomal subunit protein uS11A / Small ribosomal subunit protein uS8A / Small ribosomal subunit protein uS12A / Small ribosomal subunit protein eS24A / Small ribosomal subunit protein eS4A / Small ribosomal subunit protein eS6A / Small ribosomal subunit protein eS8A / Small ribosomal subunit protein uS17A ...Small ribosomal subunit protein uS4A / Small ribosomal subunit protein uS15 / Small ribosomal subunit protein uS11A / Small ribosomal subunit protein uS8A / Small ribosomal subunit protein uS12A / Small ribosomal subunit protein eS24A / Small ribosomal subunit protein eS4A / Small ribosomal subunit protein eS6A / Small ribosomal subunit protein eS8A / Small ribosomal subunit protein uS17A / Small ribosomal subunit protein uS9A / Small ribosomal subunit protein uS13A / rRNA 2'-O-methyltransferase fibrillarin / Exosome complex component RRP43 / Ribosomal RNA-processing protein 7 / KRR1 small subunit processome component / Periodic tryptophan protein 2 / Small ribosomal subunit protein uS7 / Small ribosomal subunit protein eS7A / U3 small nucleolar ribonucleoprotein protein IMP3 / Small ribosomal subunit protein eS1A / Protein SOF1 / U3 small nucleolar RNA-associated protein 11 / U3 small nucleolar RNA-associated protein 20 / Small ribosomal subunit protein eS27A / Ribosome biogenesis protein UTP30 / Essential nuclear protein 1 / Exosome complex component RRP4 / U3 small nucleolar RNA-associated protein 9 / 13 kDa ribonucleoprotein-associated protein / U3 small nucleolar RNA-associated protein 7 / U3 small nucleolar RNA-associated protein 18 / Regulator of rDNA transcription protein 14 / Protein FAF1 / U3 small nucleolar RNA-associated protein 10 / Exosome complex component SKI6 / ATP-dependent RNA helicase DOB1 / U3 small nucleolar RNA-associated protein MPP10 / Ribosome biogenesis protein ENP2 / Exosome complex component MTR3 / U3 small nucleolar RNA-associated protein 22 / Exosome complex component RRP46 / U3 small nucleolar RNA-associated protein 8 / Exosome complex component CSL4 / RNA cytidine acetyltransferase / U3 small nucleolar ribonucleoprotein protein IMP4 / U3 small nucleolar RNA-associated protein 6 / NET1-associated nuclear protein 1 / U3 small nucleolar RNA-associated protein 5 / U3 small nucleolar RNA-associated protein 15 / U3 small nucleolar RNA-associated protein 14 / rRNA biogenesis protein RRP5 / rRNA-processing protein FCF1 / Exosome complex component RRP45 / U3 small nucleolar RNA-associated protein 13 / U3 small nucleolar RNA-associated protein 21 / Ribosomal RNA small subunit methyltransferase NEP1 / Ribosomal RNA-processing protein 9 / Nucleolar complex protein 4 / Protein BFR2 / U3 small nucleolar RNA-associated protein 4 / RNA 3'-terminal phosphate cyclase-like protein / Exosome complex exonuclease DIS3 / Exosome complex component RRP40 / U3 small nucleolar RNA-associated protein 16 / Ribosome biogenesis protein BMS1 / rRNA-processing protein FCF2 / Something about silencing protein 10 / U3 small nucleolar RNA-associated protein 12 / Exosome complex component RRP42 / Nucleolar protein 56 / Nucleolar protein 58 / Small ribosomal subunit protein eS28A / Nucleolar complex protein 14 / Pre-rRNA-processing protein PNO1 Similarity search - Component | |||||||||||||||
| Biological species |    | |||||||||||||||
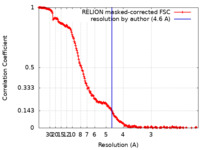
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||||||||
 Authors Authors | Cheng J / Lau B / Flemming D / Venuta GL / Berninghausen O / Beckmann R / Hurt E | |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome. Authors: Benjamin Lau / Jingdong Cheng / Dirk Flemming / Giuseppe La Venuta / Otto Berninghausen / Roland Beckmann / Ed Hurt /  Abstract: Ribosome assembly is catalyzed by numerous trans-acting factors and coupled with irreversible pre-rRNA processing, driving the pathway toward mature ribosomal subunits. One decisive step early in ...Ribosome assembly is catalyzed by numerous trans-acting factors and coupled with irreversible pre-rRNA processing, driving the pathway toward mature ribosomal subunits. One decisive step early in this progression is removal of the 5' external transcribed spacer (5'-ETS), an RNA extension at the 18S rRNA that is integrated into the huge 90S pre-ribosome structure. Upon endo-nucleolytic cleavage at an internal site, A, the 5'-ETS is separated from the 18S rRNA and degraded. Here we present biochemical and cryo-electron microscopy analyses that depict the RNA exosome, a major 3'-5' exoribonuclease complex, in a super-complex with the 90S pre-ribosome. The exosome is docked to the 90S through its co-factor Mtr4 helicase, a processive RNA duplex-dismantling helicase, which strategically positions the exosome at the base of 5'-ETS helices H9-H9', which are dislodged in our 90S-exosome structures. These findings suggest a direct role of the exosome in structural remodeling of the 90S pre-ribosome to drive eukaryotic ribosome synthesis. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11807.map.gz emd_11807.map.gz | 332.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11807-v30.xml emd-11807-v30.xml emd-11807.xml emd-11807.xml | 115.6 KB 115.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11807_fsc.xml emd_11807_fsc.xml | 19.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11807.png emd_11807.png | 170 KB | ||
| Others |  emd_11807_additional_1.map.gz emd_11807_additional_1.map.gz | 486.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11807 http://ftp.pdbj.org/pub/emdb/structures/EMD-11807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11807 | HTTPS FTP |
-Validation report
| Summary document |  emd_11807_validation.pdf.gz emd_11807_validation.pdf.gz | 266.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11807_full_validation.pdf.gz emd_11807_full_validation.pdf.gz | 265.3 KB | Display | |
| Data in XML |  emd_11807_validation.xml.gz emd_11807_validation.xml.gz | 16.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11807 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11807 | HTTPS FTP |
-Related structure data
| Related structure data |  7ajtMC  7ajuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11807.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11807.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Map without post processing
| File | emd_11807_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map without post processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 90S-exosome super-complex in state Pre-A1-exosome
| Entire | Name: 90S-exosome super-complex in state Pre-A1-exosome |
|---|---|
| Components |
|
+Supramolecule #1: 90S-exosome super-complex in state Pre-A1-exosome
| Supramolecule | Name: 90S-exosome super-complex in state Pre-A1-exosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#79 |
|---|---|
| Source (natural) | Organism:  |
+Macromolecule #1: rRNA 2'-O-methyltransferase fibrillarin
| Macromolecule | Name: rRNA 2'-O-methyltransferase fibrillarin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.525418 KDa |
| Sequence | String: MSFRPGSRGG SRGGSRGGFG GRGGSRGGAR GGSRGGFGGR GGSRGGARGG SRGGFGGRGG SRGGARGGSR GGRGGAAGGA RGGAKVVIE PHRHAGVYIA RGKEDLLVTK NMAPGESVYG EKRISVEEPS KEDGVPPTKV EYRVWNPFRS KLAAGIMGGL D ELFIAPGK ...String: MSFRPGSRGG SRGGSRGGFG GRGGSRGGAR GGSRGGFGGR GGSRGGARGG SRGGFGGRGG SRGGARGGSR GGRGGAAGGA RGGAKVVIE PHRHAGVYIA RGKEDLLVTK NMAPGESVYG EKRISVEEPS KEDGVPPTKV EYRVWNPFRS KLAAGIMGGL D ELFIAPGK KVLYLGAASG TSVSHVSDVV GPEGVVYAVE FSHRPGRELI SMAKKRPNII PIIEDARHPQ KYRMLIGMVD CV FADVAQP DQARIIALNS HMFLKDQGGV VISIKANCID STVDAETVFA REVQKLREER IKPLEQLTLE PYERDHCIVV GRY MRSGLK K |
+Macromolecule #2: 40S ribosomal protein S1-A
| Macromolecule | Name: 40S ribosomal protein S1-A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.798467 KDa |
| Sequence | String: MAVGKNKRLS KGKKGQKKRV VDPFTRKEWF DIKAPSTFEN RNVGKTLVNK STGLKSASDA LKGRVVEVCL ADLQGSEDHS FRKIKLRVD EVQGKNLLTN FHGMDFTTDK LRSMVRKWQT LIEANVTVKT SDDYVLRIFA IAFTRKQANQ VKRHSYAQSS H IRAIRKVI ...String: MAVGKNKRLS KGKKGQKKRV VDPFTRKEWF DIKAPSTFEN RNVGKTLVNK STGLKSASDA LKGRVVEVCL ADLQGSEDHS FRKIKLRVD EVQGKNLLTN FHGMDFTTDK LRSMVRKWQT LIEANVTVKT SDDYVLRIFA IAFTRKQANQ VKRHSYAQSS H IRAIRKVI SEILTKEVQG STLAQLTSKL IPEVINKEIE NATKDIFPLQ NIHVRKVKLL KQPKFDVGAL MALHGEGSGE EK GKKVTGF KDEVLETV |
+Macromolecule #3: RNA cytidine acetyltransferase
| Macromolecule | Name: RNA cytidine acetyltransferase / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 119.509445 KDa |
| Sequence | String: MAKKAIDSRI PSLIRNGVQT KQRSIFVIVG DRARNQLPNL HYLMMSADLK MNKSVLWAYK KKLLGFTSHR KKRENKIKKE IKRGTREVN EMDPFESFIS NQNIRYVYYK ESEKILGNTY GMCILQDFEA LTPNLLARTI ETVEGGGIVV ILLKSMSSLK Q LYTMTMDV ...String: MAKKAIDSRI PSLIRNGVQT KQRSIFVIVG DRARNQLPNL HYLMMSADLK MNKSVLWAYK KKLLGFTSHR KKRENKIKKE IKRGTREVN EMDPFESFIS NQNIRYVYYK ESEKILGNTY GMCILQDFEA LTPNLLARTI ETVEGGGIVV ILLKSMSSLK Q LYTMTMDV HARYRTEAHG DVVARFNERF ILSLGSNPNC LVVDDELNVL PLSGAKNVKP LPPKEDDELP PKQLELQELK ES LEDVQPA GSLVSLSKTV NQAHAILSFI DAISEKTLNF TVALTAGRGR GKSAALGISI AAAVSHGYSN IFVTSPSPEN LKT LFEFIF KGFDALGYQE HIDYDIIQST NPDFNKAIVR VDIKRDHRQT IQYIVPQDHQ VLGQAELVVI DEAAAIPLPI VKNL LGPYL VFMASTINGY EGTGRSLSLK LIQQLRNQNN TSGRESTQTA VVSRDNKEKD SHLHSQSRQL REISLDEPIR YAPGD PIEK WLNKLLCLDV TLIKNPRFAT RGTPHPSQCN LFVVNRDTLF SYHPVSENFL EKMMALYVSS HYKNSPNDLQ LMSDAP AHK LFVLLPPIDP KDGGRIPDPL CVIQIALEGE ISKESVRNSL SRGQRAGGDL IPWLISQQFQ DEEFASLSGA RIVRIAT NP EYASMGYGSR AIELLRDYFE GKFTDMSEDV RPKDYSIKRV SDKELAKTNL LKDDVKLRDA KTLPPLLLKL SEQPPHYL H YLGVSYGLTQ SLHKFWKNNS FVPVYLRQTA NDLTGEHTCV MLNVLEGRES NWLVEFAKDF RKRFLSLLSY DFHKFTAVQ ALSVIESSKK AQDLSDDEKH DNKELTRTHL DDIFSPFDLK RLDSYSNNLL DYHVIGDMIP MLALLYFGDK MGDSVKLSSV QSAILLAIG LQRKNIDTIA KELNLPSNQT IAMFAKIMRK MSQYFRQLLS QSIEETLPNI KDDAIAEMDG EEIKNYNAAE A LDQMEEDL EEAGSEAVQA MREKQKELIN SLNLDKYAIN DNSEEWAESQ KSLEIAAKAK GVVSLKTGKK RTTEKAEDIY RQ EMKAMKK PRKSKKAAN |
+Macromolecule #4: Periodic tryptophan protein 2
| Macromolecule | Name: Periodic tryptophan protein 2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.097039 KDa |
| Sequence | String: MKSDFKFSNL LGTVYRQGNI TFSDDGKQLL SPVGNRVSVF DLINNKSFTF EYEHRKNIAA IDLNKQGTLL ISIDEDGRAI LVNFKARNV LHHFNFKEKC SAVKFSPDGR LFALASGRFL QIWKTPDVNK DRQFAPFVRH RVHAGHFQDI TSLTWSQDSR F ILTTSKDL ...String: MKSDFKFSNL LGTVYRQGNI TFSDDGKQLL SPVGNRVSVF DLINNKSFTF EYEHRKNIAA IDLNKQGTLL ISIDEDGRAI LVNFKARNV LHHFNFKEKC SAVKFSPDGR LFALASGRFL QIWKTPDVNK DRQFAPFVRH RVHAGHFQDI TSLTWSQDSR F ILTTSKDL SAKIWSVDSE EKNLAATTFN GHRDYVMGAF FSHDQEKIYT VSKDGAVFVW EFTKRPSDDD DNESEDDDKQ EE VDISKYS WRITKKHFFY ANQAKVKCVT FHPATRLLAV GFTSGEFRLY DLPDFTLIQQ LSMGQNPVNT VSVNQTGEWL AFG SSKLGQ LLVYEWQSES YILKQQGHFD STNSLAYSPD GSRVVTASED GKIKVWDITS GFCLATFEEH TSSVTAVQFA KRGQ VMFSS SLDGTVRAWD LIRYRNFRTF TGTERIQFNC LAVDPSGEVV CAGSLDNFDI HVWSVQTGQL LDALSGHEGP VSCLS FSQE NSVLASASWD KTIRIWSIFG RSQQVEPIEV YSDVLALSMR PDGKEVAVST LKGQISIFNI EDAKQVGNID CRKDII SGR FNQDRFTAKN SERSKFFTTI HYSFDGMAIV AGGNNNSICL YDVPNEVLLK RFIVSRNMAL NGTLEFLNSK KMTEAGS LD LIDDAGENSD LEDRIDNSLP GSQRGGDLST RKMRPEVRVT SVQFSPTANA FAAASTEGLL IYSTNDTILF DPFDLDVD V TPHSTVEALR EKQFLNALVM AFRLNEEYLI NKVYEAIPIK EIPLVASNIP AIYLPRILKF IGDFAIESQH IEFNLIWIK ALLSASGGYI NEHKYLFSTA MRSIQRFIVR VAKEVVNTTT DNKYTYRFLV STDGSMEDGA ADDDEVLLKD DADEDNEENE ENDVVMESD DEEGWIGFNG KDNKLPLSNE NDSSDEEENE KELP |
+Macromolecule #5: Nucleolar complex protein 14
| Macromolecule | Name: Nucleolar complex protein 14 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 94.463195 KDa |
| Sequence | String: MAGSQLKNLK AALKARGLTG QTNVKSKNKK NSKRQAKEYD REEKKKAIAE IREEFNPFEI KAARNKRRDG LPSKTADRIA VGKPGISKQ IGEEQRKRAF EARKMMKNKR GGVIDKRFGE RDKLLTEEEK MLERFTRERQ SQSKRNANLF NLEDDEDDGD M FGDGLTHL ...String: MAGSQLKNLK AALKARGLTG QTNVKSKNKK NSKRQAKEYD REEKKKAIAE IREEFNPFEI KAARNKRRDG LPSKTADRIA VGKPGISKQ IGEEQRKRAF EARKMMKNKR GGVIDKRFGE RDKLLTEEEK MLERFTRERQ SQSKRNANLF NLEDDEDDGD M FGDGLTHL GQSLSLEDEL ANDEEDFLAS KRFNEDDAEL QQPQRKKTKA EVMKEVIAKS KFYKQERQKA QGIMEDQIDN LD DNFEDVM SELMMTQPKK NPMEPKTDLD KEYDIKVKEL QLDKRAAPSD RTKTEEEKNA EAEEKKRELE QQRLDRMNGM IEL EEGEER GVEDLDDGFW ENEEDYEDDN DGIADSDDDI KFEDQGRDEG FSQILKKKNI SISCPRTHDA LLDQVKKLDL DDHP KIVKN IIKAYQPKLA EGNKEKLGKF TAVLLRHIIF LSNQNYLKNV QSFKRTQNAL ISILKSLSEK YNRELSEECR DYINE MQAR YKKNHFDALS NGDLVFFSII GILFSTSDQY HLVITPALIL MSQFLEQIKF NSLKRIAFGA VLVRIVSQYQ RISKRY IPE VVYFFQKILL TFIVEKENQE KPLDFENIRL DSYELGLPLD VDFTKKRSTI IPLHTLSTMD TEAHPVDQCV SVLLNVM ES LDATISTVWK SLPAFNEIIL PIQQLLSAYT SKYSDFEKPR NILNKVEKLT KFTEHIPLAL QNHKPVSIPT HAPKYEEN F NPDKKSYDPD RTRSEINKMK AQLKKERKFT MKEIRKDAKF EARQRIEEKN KESSDYHAKM AHIVNTINTE EGAEKNKYE RERKLRGGKK |
+Macromolecule #6: Something about silencing protein 10
| Macromolecule | Name: Something about silencing protein 10 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 70.364398 KDa |
| Sequence | String: MVRKGSNRTK TSEVGDEINP YGLNEVDDFA SKREKVLLGQ STFGDSNKDD DHSLLEDEDE EEVLAMDEDD ESIDEREDEE EEEEEELDG AAAYKKIFGR NLETDQLPEE DEENGMLDNE NAWGSTKGEY YGADDLDDDE AAKEIEKEAL RQQKKHLEEL N MNDYLDEE ...String: MVRKGSNRTK TSEVGDEINP YGLNEVDDFA SKREKVLLGQ STFGDSNKDD DHSLLEDEDE EEVLAMDEDD ESIDEREDEE EEEEEELDG AAAYKKIFGR NLETDQLPEE DEENGMLDNE NAWGSTKGEY YGADDLDDDE AAKEIEKEAL RQQKKHLEEL N MNDYLDEE EEEEWVKSAK EFDMGEFKNS TKQADTKTSI TDILNMDDEA RDNYLRTMFP EFAPLSKEFT ELAPKFDELK KS EENEFNK LKLIALGSYL GTISCYYSIL LHELHNNEDF TSMKGHPVME KILTTKEIWR QASELPSSFD VNEGDGSESE ETA NIEAFN EKKLNELQNS EDSDAEDGGK QKQEIDEEER ESDEEEEEED VDIDDFEEYV AQSRLHSKPK TSSMPEADDF IESE IADVD AQDKKARRRT LRFYTSKIDQ QENKKTDRFK GDDDIPYKER LFERQQRLLD EARKRGMHDN NGADLDDKDY GSEDE AVSR SINTQGENDY YQQVQRGKQD KKISRKEAHK NAVIAAREGK LAELAENVSG DGKRAINYQI LKNKGLTPKR NKDNRN SRV KKRKKYQKAQ KKLKSVRAVY SGGQSGVYEG EKTGIKKGLT RSVKFKN |
+Macromolecule #7: U3 small nucleolar RNA-associated protein 4
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 4 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.909242 KDa |
| Sequence | String: MSSSLLSVLK EKSRSLKIRN KPVKMTSQER MIVHRCRFVD FTPATITSLA FSHKSNINKL TPSDLRLAIG RSNGNIEIWN PRNNWFQEM VIEGGKDRSI EGLCWSNVNG ESLRLFSIGG STVVTEWDLA TGLPLRNYDC NSGVIWSISI NDSQDKLSVG C DNGTVVLI ...String: MSSSLLSVLK EKSRSLKIRN KPVKMTSQER MIVHRCRFVD FTPATITSLA FSHKSNINKL TPSDLRLAIG RSNGNIEIWN PRNNWFQEM VIEGGKDRSI EGLCWSNVNG ESLRLFSIGG STVVTEWDLA TGLPLRNYDC NSGVIWSISI NDSQDKLSVG C DNGTVVLI DISGGPGVLE HDTILMRQEA RVLTLAWKKD DFVIGGCSDG RIRIWSAQKN DENMGRLLHT MKVDKAKKES TL VWSVIYL PRTDQIASGD STGSIKFWDF QFATLNQSFK AHDADVLCLT TDTDNNYVFS AGVDRKIFQF SQNTNKSQKN NRW VNSSNR LLHGNDIRAI CAYQSKGADF LVSGGVEKTL VINSLTSFSN GNYRKMPTVE PYSKNVLVNK EQRLVVSWSE STVK IWTMG TDSSTEQNYK LVCKLTLKDD QNISTCSLSP DGQVLVVGRP STTKVFHLQP VGNKLKVTKL DNDLLLRTST KLVKF IDNS KIVICSCEDD VFIVDLESEE DEKPQEVELL EVTSTKSSIK VPYINRINHL EVDQNIAVIS RGCGVVDILD LKARIS KPL ARLNNFITAV HINTSRKSVV VITADNKIYE FNMNLNSEAE NEDSESVLTQ WSKNNTDNLP KEWKTLKENC VGIFSDI EN SSRLWFWGAT WISRIDFDVD FPINKRRKQK KRTHEGLTIT DESNFMNDEE DDEDDDIDME ISENLNVLLN QGNKIKST D VQRNEESSGH FFFTDKYKPL LFVDLISSNE LAIIERNPLT FHSKQKAFIQ PKLVF |
+Macromolecule #8: U3 small nucleolar RNA-associated protein 5
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 5 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.079445 KDa |
| Sequence | String: MDSPVLQSAY DPSGQYLCYV TVALDKQRVG VQPTQRATSS GVDTVWNENF LYLEDSKLKV TCLKWVNLAS SDTVAIILGM NNGEIWLYS VLANEVTYKF TTGNSYEIKD IDLMGNQLWC IDSSDAFYQF DLLQFKLLQH FRINNCVQLN KLTIVPAGDS V AQLLVASH ...String: MDSPVLQSAY DPSGQYLCYV TVALDKQRVG VQPTQRATSS GVDTVWNENF LYLEDSKLKV TCLKWVNLAS SDTVAIILGM NNGEIWLYS VLANEVTYKF TTGNSYEIKD IDLMGNQLWC IDSSDAFYQF DLLQFKLLQH FRINNCVQLN KLTIVPAGDS V AQLLVASH SISLIDIEEK KVVMTFPGHV SPVSTLQVIT NEFFISGAEG DRFLNVYDIH SGMTKCVLVA ESDIKELSHS GQ ADSIAVT TEDGSLEIFV DPLVSSSTKK RGNKSKKSSK KIQIVSKDGR KVPIYNAFIN KDLLNVSWLQ NATMPYFKNL QWR EIPNEY TVEISLNWNN KNKSADRDLH GKDLASATNY VEGNARVTSG DNFKHVDDAI KSWERELTSL EQEQAKPPQA NELL TETFG DKLESSTVAR ISGKKTNLKG SNLKTATTTG TVTVILSQAL QSNDHSLLET VLNNRDERVI RDTIFRLKPA LAVIL LERL AERIARQTHR QGPLNVWVKW CLIIHGGYLV SIPNLMSTLS SLHSTLKRRS DLLPRLLALD ARLDCTINKF KTLNYE AGD IHSSEPVVEE DEDDVEYNEE LDDAGLIEDG EESYGSEEEE EGDSDNEEEQ KHTSSKQDGR LETEQSDGEE EAGYSDV EM E |
+Macromolecule #9: U3 small nucleolar RNA-associated protein 6
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 6 / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.495277 KDa |
| Sequence | String: MSKTRYYLEQ CIPEMDDLVE KGLFTKNEVS LIMKKRTDFE HRLNSRGSSI NDYIKYINYE SNVNKLRAKR CKRILQVKKT NSLSDWSIQ QRIGFIYQRG TNKFPQDLKF WAMYLNYMKA RGNQTSYKKI HNIYNQLLKL HPTNVDIWIS CAKYEYEVHA N FKSCRNIF ...String: MSKTRYYLEQ CIPEMDDLVE KGLFTKNEVS LIMKKRTDFE HRLNSRGSSI NDYIKYINYE SNVNKLRAKR CKRILQVKKT NSLSDWSIQ QRIGFIYQRG TNKFPQDLKF WAMYLNYMKA RGNQTSYKKI HNIYNQLLKL HPTNVDIWIS CAKYEYEVHA N FKSCRNIF QNGLRFNPDV PKLWYEYVKF ELNFITKLIN RRKVMGLINE REQELDMQNE QKNNQAPDEE KSHLQVPSTG DS MKDKLNE LPEADISVLG NAETNPALRG DIALTIFDVC MKTLGKHYIN KHKGYYAISD SKMNIELNKE TLNYLFSESL RYI KLFDEF LDLERDYLIN HVLQFWKNDM YDLSLRKDLP ELYLKTVMID ITLNIRYMPV EKLDIDQLQL SVKKYFAYIS KLDS ASVKS LKNEYRSYLQ DNYLKKMNAE DDPRYKILDL IISKL |
+Macromolecule #10: U3 small nucleolar RNA-associated protein 7
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 7 / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.41857 KDa |
| Sequence | String: MGHKKNGHRR QIKERENQNK FERSTYTNNA KNNHTQTKDK KLRAGLKKID EQYKKAVSSA AATDYLLPES NGYLEPENEL EKTFKVQQS EIKSSVDVST ANKALDLSLK EFGPYHIKYA KNGTHLLITG RKGHVASMDW RKGQLRAELF LNETCHSATY L QNEQYFAV ...String: MGHKKNGHRR QIKERENQNK FERSTYTNNA KNNHTQTKDK KLRAGLKKID EQYKKAVSSA AATDYLLPES NGYLEPENEL EKTFKVQQS EIKSSVDVST ANKALDLSLK EFGPYHIKYA KNGTHLLITG RKGHVASMDW RKGQLRAELF LNETCHSATY L QNEQYFAV AQKKYTFIYD HEGTELHRLK QHIEARHLDF LPYHYLLVTA GETGWLKYHD VSTGQLVSEL RTKAGPTMAM AQ NPWNAVM HLGHSNGTVS LWSPSMPEPL VKLLSARGPV NSIAIDRSGY YMATTGADRS MKIWDIRNFK QLHSVESLPT PGT NVSISD TGLLALSRGP HVTLWKDALK LSGDSKPCFG SMGGNPHRNT PYMSHLFAGN KVENLGFVPF EDLLGVGHQT GITN LIVPG AGEANYDALE LNPFETKKQR QEQEVRTLLN KLPADTITLD PNSIGSVDKR SSTIRLNAKD LAQTTMDANN KAKTN SDIP DVKPDVKGKN SGLRSFLRKK TQNVIDERKL RVQKQLDKEK NIRKRNHQIK QGLISEDHKD VIEEALSRFG |
+Macromolecule #11: U3 small nucleolar RNA-associated protein 8
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 8 / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.269648 KDa |
| Sequence | String: MPSLSQPFRL ATLPKIASLS NFSLQADYVQ VADGTFNEST NNITLGISGS SISQYIINPT PKLTFDYPIP STNIITACNA EKGQANIDG NIEASTDDEA NNEKTINTQK KRNVEIWAFG LMVNKGNYTL NVITKALEDT TDTSNDHLSE SDIDNKAYTG S DEFLSQYK ...String: MPSLSQPFRL ATLPKIASLS NFSLQADYVQ VADGTFNEST NNITLGISGS SISQYIINPT PKLTFDYPIP STNIITACNA EKGQANIDG NIEASTDDEA NNEKTINTQK KRNVEIWAFG LMVNKGNYTL NVITKALEDT TDTSNDHLSE SDIDNKAYTG S DEFLSQYK IKAKAKVMSI KIDTKNSLVI AILQNGLIEI FDFKLTLLHS FDISYDNLKY AKWFTENGTE YVFVLCPLQD DK VCYKLLE LTDCGSGESS PIKELSSTII EGFSFENSKL CYQFGKLYKL NQGKIYIYSL PHCQLQQVIE FPMVDKLSPG DDL ISFQPV SVNRVLLTVN NVIYLLDLLH CSTLSQRELT HVKTFQLLKS AVINSEKSHN SKTIAIGIST KNGPNPTSSL EIIN IDVGT NTLKDSLGKS FQVGNNDSSV ILKPLFDDKD INDKRVKCND VSGDSSVPVL HCNEVIEKLS ALQDNDITSF DDIFF KELK IKEEHYTEKD RYISDPGFLN KVLDLIFGKF SGNDYPKTLT FLLTHPLFPL SRTRNLLSLL RDQPRLFKQA IVTCPN LPL NELLEELFSI RNRELLLDIS FRILQDFTRD SIKQEMKKLS KLDVQNFIEF ITSGGEDSSP ECFNPSQSTQ LFQLLSL VL DSIGLFSLEG ALLENLTLYI DKQVEIAERN TELWNLIDTK GFQHGFASST FDNGTSQKRA LPTYTMEYLD I |
+Macromolecule #12: U3 small nucleolar RNA-associated protein 9
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 9 / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.347254 KDa |
| Sequence | String: MGSSLDLVAS FSHDSTRFAF QASVAQKNNV DIYPLNETKD YVVNSSLVSH IDYETNDMKV SDVIFFGWCS DLIDTQSSNI KRKLDEDEG TGESSEQRCE NFFVNGFPDG RIVVYSSNGK DIVNIIKNKK EILGADTDES DIWILDSDKV VKKLQYNNSK P LKTFTLVD ...String: MGSSLDLVAS FSHDSTRFAF QASVAQKNNV DIYPLNETKD YVVNSSLVSH IDYETNDMKV SDVIFFGWCS DLIDTQSSNI KRKLDEDEG TGESSEQRCE NFFVNGFPDG RIVVYSSNGK DIVNIIKNKK EILGADTDES DIWILDSDKV VKKLQYNNSK P LKTFTLVD GKDDEIVHFQ ILHQNGTLLV CIITKQMVYI VDPSKRRPST KYSFEISDAV ACEFSSDGKY LLIANNEELI AY DLKEDSK LIQSWPVQVK TLKTLDDLIM ALTTDGKINN YKIGEADKVC SIVVNEDLEI IDFTPINSKQ QVLISWLNVN EPN FESISL KEIETQGYIT INKNEKNNAD EADQKKLEEK EEEAQPEVQH EKKETETKIN KKVSKSDQVE IANILSSHLE ANST EILDD LMSGSWTEPE IKKFILTKIN TVDHLSKIFL TISKSITQNP WNEENLLPLW LKWLLTLKSG ELNSIKDKHT KKNCK HLKS ALRSSEEILP VLLGIQGRLE MLRRQAKLRE DLAQLSMQEG EDDEIEVIEH SNVISNPLQD QASPVEKLEP DSIVYA NGE SDEFVDASEY KD |
+Macromolecule #13: U3 small nucleolar RNA-associated protein 10
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 10 / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200.298984 KDa |
| Sequence | String: MSSLSDQLAQ VASNNATVAL DRKRRQKLHS ASLIYNSKTA ATQDYDFIFE NASKALEELS QIEPKFAIFS RTLFSESSIS LDRNVQTKE EIKDLDNAIN AYLLLASSKW YLAPTLHATE WLVRRFQIHV KNTEMLLLST LNYYQTPVFK RILSIIKLPP L FNCLSNFV ...String: MSSLSDQLAQ VASNNATVAL DRKRRQKLHS ASLIYNSKTA ATQDYDFIFE NASKALEELS QIEPKFAIFS RTLFSESSIS LDRNVQTKE EIKDLDNAIN AYLLLASSKW YLAPTLHATE WLVRRFQIHV KNTEMLLLST LNYYQTPVFK RILSIIKLPP L FNCLSNFV RSEKPPTALT MIKLFNDMDF LKLYTSYLDQ CIKHNATYTN QLLFTTCCFI NVVAFNSNND EKLNQLVPIL LE ISAKLLA SKSKDCQIAA HTILVVFATA LPLKKTIILA AMETILSNLD AKEAKHSALL TICKLFQTLK GQGNVDQLPS KIF KLFDSK FDTVSILTFL DKEDKPVCDK FITSYTRSIA RYDRSKLNII LSLLKKIRLE RYEVRLIITD LIYLSEILED KSQL VELFE YFISINEDLV LKCLKSLGLT GELFEIRLTT SLFTNADVNT DIVKQLSDPV ETTKKDTASF QTFLDKHSEL INTTN VSML TETGERYKKV LSLFTEAIGK GYKASSFLTS FFTTLESRIT FLLRVTISPA APTALKLISL NNIAKYINSI EKEVNI FTL VPCLICALRD ASIKVRTGVK KILSLIAKRP STKHYFLSDK LYGENVTIPM LNPKDSEAWL SGFLNEYVTE NYDISRI LT PKRNEKVFLM FWANQALLIP SPYAKTVLLD NLNKSPTYAS SYSSLFEEFI SHYLENRSSW EKSCIANKTN FEHFERSL V NLVSPKEKQS FMIDFVLSAL NSDYEQLANI AAERLISIFA SLNNAQKLKI VQNIVDSSSN VESSYDTVGV LQSLPLDSD IFVSILNQNS ISNEMDQTDF SKRRRRRSST SKNAFLKEEV SQLAELHLRK LTIILEALDK VRNVGSEKLL FTLLSLLSDL ETLDQDGGL PVLYAQETLI SCTLNTITYL KEHGCTELTN VRADILVSAI RNSASPQVQN KLLLVIGSLA TLSSEVILHS V MPIFTFMG AHSIRQDDEF TTKVVERTIL TVVPALIKNS KGNEKEEMEF LLLSFTTALQ HVPRHRRVKL FSTLIKTLDP VK ALGSFLF LIAQQYSSAL VNFKIGEARI LIEFIKALLV DLHVNEELSG LNDLLDIIKL LTSSKSSSEK KKSLESRVLF SNG VLNFSE SEFLTFMNNT FEFINKITEE TDQDYYDVRR NLRLKVYSVL LDETSDKKLI RNIREEFGTL LEGVLFFINS VELT FSCIT SQENEEASDS ETSLSDHTTE IKEILFKVLG NVLQILPVDE FVNAVLPLLS TSTNEDIRYH LTLVIGSKFE LEGSE AIPI VNNVMKVLLD RMPLESKSVV ISQVILNTMT ALVSKYGKKL EGSILTQALT LATEKVSSDM TEVKISSLAL ITNCVQ VLG VKSIAFYPKI VPPSIKLFDA SLADSSNPLK EQLQVAILLL FAGLIKRIPS FLMSNILDVL HVIYFSREVD SSIRLSV IS LIIENIDLKE VLKVLFRIWS TEIATSNDTV AVSLFLSTLE STVENIDKKS ATSQSPIFFK LLLSLFEFRS ISSFDNNT I SRIEASVHEI SNSYVLKMND KVFRPLFVIL VRWAFDGEGV TNAGITETER LLAFFKFFNK LQENLRGIIT SYFTYLLEP VDMLLKRFIS KDMENVNLRR LVINSLTSSL KFDRDEYWKS TSRFELISVS LVNQLSNIEN SIGKYLVKAI GALASNNSGV DEHNQILNK LIVEHMKASC SSNEKLWAIR AMKLIYSKIG ESWLVLLPQL VPVIAELLED DDEEIEREVR TGLVKVVENV L GEPFDRYL D |
+Macromolecule #14: U3 small nucleolar RNA-associated protein 11
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 11 / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.806348 KDa |
| Sequence | String: MAKLVHDVQK KQHRERSQLT SRSRYGFLEK HKDYVKRAQD FHRKQSTLKV LREKAKERNP DEYYHAMHSR KTDAKGLLIS SRHGDEEDE SLSMDQVKLL KTQDSNYVRT LRQIELKKLE KGAKQLMFKS SGNHTIFVDS REKMNEFTPE KFFNTTSEMV N RSENRLTK ...String: MAKLVHDVQK KQHRERSQLT SRSRYGFLEK HKDYVKRAQD FHRKQSTLKV LREKAKERNP DEYYHAMHSR KTDAKGLLIS SRHGDEEDE SLSMDQVKLL KTQDSNYVRT LRQIELKKLE KGAKQLMFKS SGNHTIFVDS REKMNEFTPE KFFNTTSEMV N RSENRLTK DQLAQDISNN RNASSIMPKE SLDKKKLKKF KQVKQHLQRE TQLKQVQQRM DAQRELLKKG SKKKIVDSSG KI SFKWKKQ RKR |
+Macromolecule #15: U3 small nucleolar RNA-associated protein 12
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 12 / type: protein_or_peptide / ID: 15 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 106.481133 KDa |
| Sequence | String: MVKSYQRFEQ AAAFGVIASN ANCVWIPASS GNSNGSGPGQ LITSALEDVN IWDIKTGDLV SKLSDGLPPG ASDARGAKPA ECTYLEAHK DTDLLAVGYA DGVIKVWDLM SKTVLLNFNG HKAAITLLQF DGTGTRLISG SKDSNIIVWD LVGEVGLYKL R SHKDSITG ...String: MVKSYQRFEQ AAAFGVIASN ANCVWIPASS GNSNGSGPGQ LITSALEDVN IWDIKTGDLV SKLSDGLPPG ASDARGAKPA ECTYLEAHK DTDLLAVGYA DGVIKVWDLM SKTVLLNFNG HKAAITLLQF DGTGTRLISG SKDSNIIVWD LVGEVGLYKL R SHKDSITG FWCQGEDWLI STSKDGMIKL WDLKTHQCIE THIAHTGECW GLAVKDDLLI TTGTDSQVKI WKLDIENDKM GG KLTEMGI FEKQSKQRGL KIEFITNSSD KTSFFYIQNA DKTIETFRIR KEEEIARGLK KREKRLKEKG LTEEEIAKSI KES YSSFIL HPFQTIRSLY KIKSASWTTV SSSKLELVLT TSSNTIEYYS IPYEKRDPTS PAPLKTHTIE LQGQRTDVRS IDIS DDNKL LATASNGSLK IWNIKTHKCI RTFECGYALT CKFLPGGLLV ILGTRNGELQ LFDLASSSLL DTIEDAHDAA IWSLD LTSD GKRLVTGSAD KTVKFWDFKV ENSLVPGTKN KFLPVLKLHH DTTLELTDDI LCVRVSPDDR YLAISLLDNT VKVFFL DSM KFYLSLYGHK LPVLSIDISF DSKMIITSSA DKNIKIWGLD FGDCHKSLFA HQDSIMNVKF LPQSHNFFSC SKDAVVK YW DGEKFECIQK LYAHQSEVWA LAVATDGGFV VSSSHDHSIR IWEETEDQVF LEEEKEKELE EQYEDTLLTS LEEGNGDD A FKADASGEGV EDEASGVHKQ TLESLKAGER LMEALDLGIA EIEGLEAYNR DMKLWQRKKL GEAPIKPQGN AVLIAVNKT PEQYIMDTLL RIRMSQLEDA LMVMPFSYVL KFLKFIDTVM QNKTLLHSHL PLICKNLFFI IKFNHKELVS QKNEELKLQI NRVKTELRS ALKSTEDDLG FNVQGLKFVK QQWNLRHNYE FVDEYDQQEK ESNSARKRVF GTVI |
+Macromolecule #16: U3 small nucleolar RNA-associated protein 13
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 13 / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 91.132562 KDa |
| Sequence | String: MDLKTSYKGI SLNPIYAGSS AVATVSENGK ILATPVLDEI NIIDLTPGSR KILHKISNED EQEITALKLT PDGQYLTYVS QAQLLKIFH LKTGKVVRSM KISSPSYILD ADSTSTLLAV GGTDGSIIVV DIENGYITHS FKGHGGTISS LKFYGQLNSK I WLLASGDT ...String: MDLKTSYKGI SLNPIYAGSS AVATVSENGK ILATPVLDEI NIIDLTPGSR KILHKISNED EQEITALKLT PDGQYLTYVS QAQLLKIFH LKTGKVVRSM KISSPSYILD ADSTSTLLAV GGTDGSIIVV DIENGYITHS FKGHGGTISS LKFYGQLNSK I WLLASGDT NGMVKVWDLV KRKCLHTLQE HTSAVRGLDI IEVPDNDEPS LNLLSGGRDD IINLWDFNMK KKCKLLKTLP VN QQVESCG FLKDGDGKRI IYTAGGDAIF QLIDSESGSV LKRTNKPIEE LFIIGVLPIL SNSQMFLVLS DQTLQLINVE EDL KNDEDT IQVTSSIAGN HGIIADMRYV GPELNKLALA TNSPSLRIIP VPDLSGPEAS LPLDVEIYEG HEDLLNSLDA TEDG LWIAT ASKDNTAIVW RYNENSCKFD IYAKYIGHSA AVTAVGLPNI VSKGYPEFLL TASNDLTIKK WIIPKPTASM DVQII KVSE YTRHAHEKDI NALSVSPNDS IFATASYDKT CKIWNLENGE LEATLANHKR GLWDVSFCQY DKLLATSSGD KTVKIW SLD TFSVMKTLEG HTNAVQRCSF INKQKQLISC GADGLIKIWD CSSGECLKTL DGHNNRLWAL STMNDGDMIV SADADGV FQ FWKDCTEQEI EEEQEKAKLQ VEQEQSLQNY MSKGDWTNAF LLAMTLDHPM RLFNVLKRAL GESRSRQDTE EGKIEVIF N EELDQAISIL NDEQLILLMK RCRDWNTNAK THTIAQRTIR CILMHHNIAK LSEIPGMVKI VDAIIPYTQR HFTRVDNLV EQSYILDYAL VEMDKLF |
+Macromolecule #17: U3 small nucleolar RNA-associated protein 14
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 14 / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 103.18975 KDa |
| Sequence | String: MAKKKSKSRS KSSRRVLDAL QLAEREINGE FDNSSDNDKR HDARRNGTVV NLLKRSKGDT NSDEDDIDSE SFEDEELNSD EALGSDDDY DILNSKFSQT IRDKKENANY QEEEDEGGYT SIDEEDLMPL SQVWDMDEKT AQSNGNDDED ASPQLKLQDT D ISSESSSS ...String: MAKKKSKSRS KSSRRVLDAL QLAEREINGE FDNSSDNDKR HDARRNGTVV NLLKRSKGDT NSDEDDIDSE SFEDEELNSD EALGSDDDY DILNSKFSQT IRDKKENANY QEEEDEGGYT SIDEEDLMPL SQVWDMDEKT AQSNGNDDED ASPQLKLQDT D ISSESSSS EESESESEDD EEEEDPFDEI SEDEEDIELN TITSKLIDET KSKAPKRLDT YGSGEANEYV LPSANAASGA SG KLSLTDM MNVIDDRQVI ENANLLKGKS STYEVPLPQR IQQRHDRKAA YEISRQEVSK WNDIVQQNRR ADHLIFPLNK PTE HNHASA FTRTQDVPQT ELQEKVDQVL QESNLANPEK DSKFEELSTA KMTPEEMRKR TTEMRLMREL MFREERKARR LKKI KSKTY RKIKKKELMK NRELAAVSSD EDNEDHDIAR AKERMTLKHK TNSKWAKDMI KHGMTNDAET REEMEEMLRQ GERLK AKML DRNSDDEEDG RVQTLSDVEN EEKENIDSEA LKSKLGKTGV MNMAFMKNGE AREREANKET LRQLRAVENG DDIKLF ESD EEETNGENIQ INKGRRIYTP GSLESNKDMN ELNDHTRKEN KVDESRSLEN RLRAKNSGQS KNARTNAEGA IIVEEES DG EPLQDGQNNQ QDEEAKDVNP WLANESDEEH TVKKQSSKVN VIDKDSSKNV KAMNKMEKAE LKQKKKKKGK SNDDEDLL L TADDSTRLKI VDPYGGSDDE QGDNVFMFKQ QDVIAEAFAG DDVVAEFQEE KKRVIDDEDD KEVDTTLPGW GEWAGAGSK PKNKKRKFIK KVKGVVNKDK RRDKNLQNVI INEKVNKKNL KYQSSAVPFP FENREQYERS LRMPIGQEWT SRASHQELIK PRIMTKPGQ VIDPLKAPFK |
+Macromolecule #18: U3 small nucleolar RNA-associated protein 15
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 15 / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.765289 KDa |
| Sequence | String: MSTARPRIIT SKAPLLPQQT TPEQRYWRQY TSAQLVKEHN SVTHISFNPQ HPHDFAVTSS TRVQIFSSRT RQVIKTFSRF KDVVYSASF RSDGKLLCAG DATGLVSVYD SYNPRTILLS INASTHPTHV TKFHTQDNKI LATASDDRVT RLWDISNAYE P QLELTGAT ...String: MSTARPRIIT SKAPLLPQQT TPEQRYWRQY TSAQLVKEHN SVTHISFNPQ HPHDFAVTSS TRVQIFSSRT RQVIKTFSRF KDVVYSASF RSDGKLLCAG DATGLVSVYD SYNPRTILLS INASTHPTHV TKFHTQDNKI LATASDDRVT RLWDISNAYE P QLELTGAT DYVRTLSFIP AAPHLVATGS YDGLIRLYDT RSSGSTPIYS LNHDQPVENV IAVSPTQIVS CGGNNFKVWD LT SNKKLYE RGNFNKAVTC LDYVENFDSP MQSALIASSL DGHVKVFDPL DNFQVKFGWK FSGPVLSCAV SPSTAQGNRH LVA GLSSGL LAIRTKKKEK RSSDKENAPA SFNKNAKSNN FQRMMRGSEY QGDQEHIIHN DKVRSQRRMR AFERNINQFK WSEA LDNAF VPGMAKELTL TVLQELRKRG KVRVALYGRD ESTLEPLLNW CLKGIEDVRS ASIVADWVAV VLELYGNTLE SSPVL QELM IDLKTKVRHE IHKSKEAQRI EGMLQLLTS |
+Macromolecule #19: Bud site selection protein 21
| Macromolecule | Name: Bud site selection protein 21 / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.431016 KDa |
| Sequence | String: MSNGHVKFDA DESQASASAV TDRQDDVLVI SKKDKEVHSS SDEESDDDDA PQEEGLHSGK SEVESQITQR EEAIRLEQSQ LRSKRRKQN ELYAKQKKSV NETEVTDEVI AELPEELLKN IDQKDEGSTQ YSSSRHVTFD KLDESDENEE ALAKAIKTKK R KTLKNLRK ...String: MSNGHVKFDA DESQASASAV TDRQDDVLVI SKKDKEVHSS SDEESDDDDA PQEEGLHSGK SEVESQITQR EEAIRLEQSQ LRSKRRKQN ELYAKQKKSV NETEVTDEVI AELPEELLKN IDQKDEGSTQ YSSSRHVTFD KLDESDENEE ALAKAIKTKK R KTLKNLRK DSVKRGKFRV QLLSTTQDSK TLPPKKESSI IRSKDRWLNR KALNKG |
+Macromolecule #20: NET1-associated nuclear protein 1
| Macromolecule | Name: NET1-associated nuclear protein 1 / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 101.341734 KDa |
| Sequence | String: MTQSLGIEQY KLSVVSGGKP ALNNLSSVTG NKNIARLSQD QRNYIIPFNN QIKVYSVETR QCVKTLKFAN NSLLSGIFLQ EEENNESIV KILLGDITVP QQEDAHLITV FTNNGHVIVL NYKGKLVESP KHFKISLADE KLANVFHSEG NYRILTTFKD P SQKAHNSL ...String: MTQSLGIEQY KLSVVSGGKP ALNNLSSVTG NKNIARLSQD QRNYIIPFNN QIKVYSVETR QCVKTLKFAN NSLLSGIFLQ EEENNESIV KILLGDITVP QQEDAHLITV FTNNGHVIVL NYKGKLVESP KHFKISLADE KLANVFHSEG NYRILTTFKD P SQKAHNSL QSYRLYALTF DDAKKQFEVA HQAEWHNVIL SNISSNGKLL AHMCKDVSTK DHEHKSISVV SLFDDSVNLS FP LGSILSS QTQSLSYNTR YVSSMAIDNM GQQLAVGFAS GVISIVSLAD LQIRLLKWHI DSVLSLSFSH DGSYLLSGGW EKV MSLWQL ETNSQQFLPR LNGIIIDCQV LGPQGNYYSL ILQMTENNSN SDYQFLLLNA SDLTSKLSIN GPLPVFNSTI KHIQ QPISA MNTKNSNSIT SLNHSKKKQS RKLIKSRRQD FTTNVEINPI NKNLYFPHIS AVQIFDFYKN EQVNYQYLTS GVNNS MGKV RFELNLQDPI ITDLKFTKDG QWMITYEIEY PPNDLLSSKD LTHILKFWTK NDNETNWNLK TKVINPHGIS VPITKI LPS PRSVNNSQGC LTADNNGGLK FWSFDSHESN WCLKKISLPN FNHFSNSVSL AWSQDGSLIF HGFDDKLQIL DFDTFKK FE SLENTKTVSE FTLDSEIQTV KLINDTNLIV ATRTTLNAIN LLRGQVINSF DLYPFVNGVY KNGHMDRLIT CDERTGNI A LVINQQLTDL DGVPTINYKS RIIIFDSDLS TKLGNFTHHE YISWIGWNYD TDFIFLDIES TLGVVGTTVN TQLSDEVNN EGILDGLVSN TITTSASNSD IFAEQLHKLS SRGKKSDTRD KNTNDNDEDE EDIALEFING EKKDKLVNMN SFTSMFDNIQ NVQMDTFFD RVMKVLT |
+Macromolecule #21: U3 small nucleolar RNA-associated protein 18
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 18 / type: protein_or_peptide / ID: 21 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.49425 KDa |
| Sequence | String: MTMATTAMNV SVPPPDEEEQ LLAKFVFGDT TDLQENLAKF NADFIFNEQE MDVEDQEDEG SESDNSEEDE AQNGELDHVN NDQLFFVDD GGNEDSQDKN EDTMDVDDED DSSSDDYSED SEEAAWIDSD DEKIKVPILV TNKTKKLRTS YNESKINGVH Y INRLRSQF ...String: MTMATTAMNV SVPPPDEEEQ LLAKFVFGDT TDLQENLAKF NADFIFNEQE MDVEDQEDEG SESDNSEEDE AQNGELDHVN NDQLFFVDD GGNEDSQDKN EDTMDVDDED DSSSDDYSED SEEAAWIDSD DEKIKVPILV TNKTKKLRTS YNESKINGVH Y INRLRSQF EKIYPRPKWV DDESDSELDD EEDDEEEGSN NVINGDINAL TKILSTTYNY KDTLSNSKLL PPKKLDIVRL KD ANASHPS HSAIQSLSFH PSKPLLLTGG YDKTLRIYHI DGKTNHLVTS LHLVGSPIQT CTFYTSLSNQ NQQNIFTAGR RRY MHSWDL SLENLTHSQT AKIEKFSRLY GHESTQRSFE NFKVAHLQNS QTNSVHGIVL LQGNNGWINI LHSTSGLWLM GCKI EGVIT DFCIDYQPIS RGKFRTILIA VNAYGEVWEF DLNKNGHVIR RWKDQGGVGI TKIQVGGGTT TTCPALQISK IKQNR WLAV GSESGFVNLY DRNNAMTSST PTPVAALDQL TTTISNLQFS PDGQILCMAS RAVKDALRLV HLPSCSVFSN WPTSGT PLG KVTSVAFSPS GGLLAVGNEQ GKVRLWKLNH Y |
+Macromolecule #22: Nucleolar complex protein 4
| Macromolecule | Name: Nucleolar complex protein 4 / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.707844 KDa |
| Sequence | String: MVLLISEIKD IAKRLTAAGD RKQYNSIIKL INELVIPENV TQLEEDETEK NLRFLVMSLF QIFRKLFSRG DLTLPSSKKS TLEKEQFVN WCRKVYEAFK TKLLAIISDI PFETSLGLDS LDVYLQLAEL ESTHFASEKG APFFPNKTFR KLIIALWSSN M GEIEDVKS ...String: MVLLISEIKD IAKRLTAAGD RKQYNSIIKL INELVIPENV TQLEEDETEK NLRFLVMSLF QIFRKLFSRG DLTLPSSKKS TLEKEQFVN WCRKVYEAFK TKLLAIISDI PFETSLGLDS LDVYLQLAEL ESTHFASEKG APFFPNKTFR KLIIALWSSN M GEIEDVKS SGASENLIIV EFTEKYYTKF ADIQYYFQSE FNQLLEDPAY QDLLLKNVGK WLALVNHDKH CSSVDADLEI FV PNPPQAI ENESKFKSNF EKNWLSLLNG QLSLQQYKSI LLILHKRIIP HFHTPTKLMD FLTDSYNLQS SNKNAGVVPI LAL NGLFEL MKRFNLEYPN FYMKLYQIIN PDLMHVKYRA RFFRLMDVFL SSTHLSAHLV ASFIKKLARL TLESPPSAIV TVIP FIYNL IRKHPNCMIM LHNPAFISNP FQTPDQVANL KTLKENYVDP FDVHESDPEL THALDSSLWE LASLMEHYHP NVATL AKIF AQPFKKLSYN MEDFLDWNYD SLLNAESSRK LKTLPTLEFE AFTNVFDNED GDSEASSQGN VYLPGVAW |
+Macromolecule #23: U3 small nucleolar RNA-associated protein 20
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 20 / type: protein_or_peptide / ID: 23 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 287.9155 KDa |
| Sequence | String: MAKQRQTTKS SKRYRYSSFK ARIDDLKIEP ARNLEKRVHD YVESSHFLAS FDQWKEINLS AKFTEFAAEI EHDVQTLPQI LYHDKKIFN SLVSFINFHD EFSLQPLLDL LAQFCHDLGP DFLKFYEEAI KTLINLLDAA IEFESSNVFE WGFNCLAYIF K YLSKFLVK ...String: MAKQRQTTKS SKRYRYSSFK ARIDDLKIEP ARNLEKRVHD YVESSHFLAS FDQWKEINLS AKFTEFAAEI EHDVQTLPQI LYHDKKIFN SLVSFINFHD EFSLQPLLDL LAQFCHDLGP DFLKFYEEAI KTLINLLDAA IEFESSNVFE WGFNCLAYIF K YLSKFLVK KLVLTCDLLI PLLSHSKEYL SRFSAEALSF LVRKCPVSNL REFVRSVFEK LEGDDEQTNL YEGLLILFTE SM TSTQETL HSKAKAIMSV LLHEALTKSS PERSVSLLSD IWMNISKYAS IESLLPVYEV MYQDFNDSLD ATNIDRILKV LTT IVFSES GRKIPDWNKI TILIERIMSQ SENCASLSQD KVAFLFALFI RNSDVKTLTL FHQKLFNYAL TNISDCFLEF FQFA LRLSY ERVFSFNGLK FLQLFLKKNW QSQGKKIALF FLEVDDKPEL QKVREVNFPE EFILSIRDFF VTAEINDSND LFEIY WRAI IFKYSKLQNT EIIIPLLERI FSTFASPDNF TKDMVGTLLK IYRKEDDASG NNLLKTILDN YENYKESLNF LRGWNK LVS NLHPSESLKG LMSHYPSLLL SLTDNFMLPD GKIRYETLEL MKTLMILQGM QVPDLLSSCM VIEEIPLTLQ NARDLTI RI KNVGAEFGKT KTDKLVSSFF LKYLFGLLTV RFSPVWTGVF DTLPNVYTKD EALVWKLVLS FIKLPDENQN LDYYQPLL E DGANKVLWDS SVVRLRDTID TFSHIWSKYS TQNTSIISTT IERRGNTTYP ILIRNQALKV MLSIPQVAEN HFVDIAPFV YNDFKTYKDE EDMENERVIT GSWTEVDRNV FLKTLSKFKN IKNVYSATEL HDHLMVLLGS RNTDVQKLAL DALLAYKNPT LNKYRDNLK NLLDDTLFKD EITTFLTENG SQSIKAEDEK VVMPYVLRIF FGRAQVPPTS GQKRSRKIAV ISVLPNFKKP Y INDFLSLA SERLDYNYFF GNSHQINSSK ATLKTIRRMT GFVNIVNSTL SVLRTNFPLH TNSVLQPLIY SIAMAYYVLD TE STEEVHL RKMASNLRQQ GLKCLSSVFE FVGNTFDWST SMEDIYAVVV KPRISHFSDE NLQQPSSLLR LFLYWAHNPS LYQ FLYYDE FATATALMDT ISNQHVKEAV IGPIIEAADS IIRNPVNDDH YVDLVTLICT SCLKILPSLY VKLSDSNSIS TFLN LLVSI TEMGFIQDDH VRSRLISSLI SILKGKLKKL QENDTQKILK ILKLIVFNYN CSWSDIEELY TTISSLFKTF DERNL RVSL TELFIELGRK VPELESISKL VADLNSYSSS RMHEYDFPRI LSTFKGLIED GYKSYSELEW LPLLFTFLHF INNKEE LAL RTNASHAIMK FIDFINEKPN LNEASKSISM LKDILLPNIR IGLRDSLEEV QSEYVSVLSY MVKNTKYFTD FEDMAIL LY NGDEEADFFT NVNHIQLHRR QRAIKRLGEH AHQLKDNSIS HYLIPMIEHY VFSDDERYRN IGNETQIAIG GLAQHMSW N QYKALLRRYI SMLKTKPNQM KQAVQLIVQL SVPLRETLRI VRDGAESKLT LSKFPSNLDE PSNFIKQELY PTLSKILGT RDDETIIERM PIAEALVNIV LGLTNDDITN FLPSILTNIC QVLRSKSEEL RDAVRVTLGK ISIILGAEYL VFVIKELMAT LKRGSQIHV LSYTVHYILK SMHGVLKHSD LDTSSSMIVK IIMENIFGFA GEEKDSENYH TKVKEIKSNK SYDAGEILAS N ISLTEFGT LLSPVKALLM VRINLRNQNK LSELLRRYLL GLNHNSDSES ESILKFCHQL FQESEMSNSP QIPKKKVKDQ VD EKEDFFL VNLESKSYTI NSNSLLLNST LQKFALDLLR NVITRHRSFL TVSHLEGFIP FLRDSLLSEN EGVVISTLRI LIT LIRLDF SDESSEIFKN CARKVLNIIK VSPSTSSELC QMGLKFLSAF IRHTDSTLKD TALSYVLGRV LPDLNEPSRQ GLAF NFLKA LVSKHIMLPE LYDIADTTRE IMVTNHSKEI RDVSRSVYYQ FLMEYDQSKG RLEKQFKFMV DNLQYPTESG RQSVM ELIN LIITKANPAL LSKLSSSFFL ALVNVSFNDD APRCREMASV LISTMLPKLE NKDLEIVEKY IAAWLKQVDN ASFLNL GLR TYKVYLKSIG FEHTIELDEL AIKRIRYILS DTSVGSEHQW DLVYSALNTF SSYMEATESV YKHGFKDIWD GIITCLL YP HSWVRQSAAN LVHQLIANKD KLEISLTNLE IQTIATRILH QLGAPSIPEN LANVSIKTLV NISILWKEQR TPFIMDVS K QTGEDLKYTT AIDYMVTRIG GIIRSDEHRM DSFMSKKACI QLLALLVQVL DEDEVIAEGE KILLPLYGYL ETYYSRAVD EEQEELRTLS NECLKILEDK LQVSDFTKIY TAVKQTVLER RKERRSKRAI LAVNAPQISA DKKLRKHARS REKRKHEKDE NGYYQRRNK RKRA |
+Macromolecule #24: U3 small nucleolar RNA-associated protein 21
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 21 / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.927844 KDa |
| Sequence | String: MSIDLKKRKV EEDVRSRGKN SKIFSPFRII GNVSNGVPFA TGTLGSTFYI VTCVGKTFQI YDANTLHLLF VSEKETPSSI VALSAHFHY VYAAYENKVG IYKRGIEEHL LELETDANVE HLCIFGDYLC ASTDDNSIFI YKKSDPQDKY PSEFYTKLTV T EIQGGEIV ...String: MSIDLKKRKV EEDVRSRGKN SKIFSPFRII GNVSNGVPFA TGTLGSTFYI VTCVGKTFQI YDANTLHLLF VSEKETPSSI VALSAHFHY VYAAYENKVG IYKRGIEEHL LELETDANVE HLCIFGDYLC ASTDDNSIFI YKKSDPQDKY PSEFYTKLTV T EIQGGEIV SLQHLATYLN KLTVVTKSNV LLFNVRTGKL VFTSNEFPDQ ITTAEPAPVL DIIALGTVTG EVIMFNMRKG KR IRTIKIP QSRISSLSFR TDGSSHLSVG TSSGDLIFYD LDRRSRIHVL KNIHRESYGG VTQATFLNGQ PIIVTSGGDN SLK EYVFDP SLSQGSGDVV VQPPRYLRSR GGHSQPPSYI AFADSQSHFM LSASKDRSLW SFSLRKDAQS QEMSQRLHKK QDGG RVGGS TIKSKFPEIV ALAIENARIG EWENIITAHK DEKFARTWDM RNKRVGRWTF DTTDDGFVKS VAMSQCGNFG FIGSS NGSI TIYNMQSGIL RKKYKLHKRA VTGISLDGMN RKMVSCGLDG IVGFYDFNKS TLLGKLKLDA PITAMVYHRS SDLFAL ALD DLSIVVIDAV TQRVVRQLWG HSNRITAFDF SPEGRWIVSA SLDSTIRTWD LPTGGCIDGI IVDNVATNVK FSPNGDL LA TTHVTGNGIC IWTNRAQFKT VSTRTIDESE FARMALPSTS VRGNDSMLSG ALESNGGEDL NDIDFNTYTS LEQIDKEL L TLSIGPRSKM NTLLHLDVIR KRSKPKEAPK KSEKLPFFLQ LSGEKVGDEA SVREGIAHET PEEIHRRDQE AQKKLDAEE QMNKFKVTGR LGFESHFTKQ LREGSQSKDY SSLLATLINF SPAAVDLEIR SLNSFEPFDE IVWFIDALTQ GLKSNKNFEL YETFMSLLF KAHGDVIHAN NKNQDIASAL QNWEDVHKKE DRLDDLVKFC MGVAAFVTTA |
+Macromolecule #25: U3 small nucleolar RNA-associated protein 22
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 22 / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.660141 KDa |
| Sequence | String: MATSVKRKAS ETSDQNIVKV QKKHSTQDST TDNGSKENDH SSQAINERTV PEQENDESDT SPESNEVATN TAATRHNGKV TATESYDIH IARETAELFK SNIFKLQIDE LLEQVKLKQK HVLKVEKFLH KLYDILQEIP DWEEKSLAEV DSFFKNKIVS V PFVDPKPI ...String: MATSVKRKAS ETSDQNIVKV QKKHSTQDST TDNGSKENDH SSQAINERTV PEQENDESDT SPESNEVATN TAATRHNGKV TATESYDIH IARETAELFK SNIFKLQIDE LLEQVKLKQK HVLKVEKFLH KLYDILQEIP DWEEKSLAEV DSFFKNKIVS V PFVDPKPI PQNTNYKFNY KKPDISLIGS FALKAGIYQP NGSSIDTLLT MPKELFEKKD FLNFRCLHKR SVYLAYLTHH LL ILLKKDK LDSFLQLEYS YFDNDPLLPI LRISCSKPTG DSLSDYNFYK TRFSINLLIG FPYKVFEPKK LLPNRNCIRI AQE SKEQSL PATPLYNFSV LSSSTHENYL KYLYKTKKQT ESFVEATVLG RLWLQQRGFS SNMSHSGSLG GFGTFEFTIL MAAL LNGGG INSNKILLHG FSSYQLFKGV IKYLATMDLC HDGHLQFHSN PENSSSSPAS KYIDEGFQTP TLFDKSTKVN ILTKM TVSS YQILKEYAGE TLRMLNNVVQ DQFSNIFLTN ISRFDNLKYD LCYDVQLPLG KYNNLETSLA ATFGSMERVK FITLEN FLA HKITNVARYA LGDRIKYIQI EMVGQKSDFP ITKRKVYSNT GGNHFNFDFV RVKLIVNPSE CDKLVTKGPA HSETMST EA AVFKNFWGIK SSLRRFKDGS ITHCCVWSTS SSEPIISSIV NFALQKHVSK KAQISNETIK KFHNFLPLPN LPSSAKTS V LNLSSFFNLK KSFDDLYKII FQMKLPLSVK SILPVGSAFR YTSLCQPVPF AYSDPDFFQD VILEFETSPK WPDEITSLE KAKTAFLLKI QEELSANSST YRSFFSRDES IPYNLEIVTL NILTPEGYGF KFRVLTERDE ILYLRAIANA RNELKPELEA TFLKFTAKY LASVRHTRTL ENISHSYQFY SPVVRLFKRW LDTHLLLGHI TDELAELIAI KPFVDPAPYF IPGSLENGFL K VLKFISQW NWKDDPLILD LVKPEDDIRD TFETSIGAGS ELDSKTMKKL SERLTLAQYK GIQMNFTNLR NSDPNGTHLQ FF VASKNDP SGILYSSGIP LPIATRLTAL AKVAVNLLQT HGLNQQTINL LFTPGLKDYD FVVDLRTPIG LKSSCGILSA TEF KNITND QAPSNFPENL NDLSEKMDPT YQLVKYLNLK YKNSLILSSR KYIGVNGGEK GDKNVITGLI KPLFKGAHKF RVNL DCNVK PVDDENVILN KEAIFHEIAA FGNDMVINFE TD |
+Macromolecule #26: rRNA-processing protein FCF1
| Macromolecule | Name: rRNA-processing protein FCF1 / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.650729 KDa |
| Sequence | String: MGKAKKTRKF GLVKRTLNTK KDQRLKKNQE NIKTKEDPEL TRNIPQVSSA LFFQYNQAIK PPYQVLIDTN FINFSIQKKV DIVRGMMDC LLAKCNPLIT DCVMAELEKL GPKYRIALKL ARDPRIKRLS CSHKGTYADD CLVHRVLQHK CYIVATNDAG L KQRIRKIP GIPLMSVGGH AYVIEKLPDV F |
+Macromolecule #27: Ribosome biogenesis protein UTP30
| Macromolecule | Name: Ribosome biogenesis protein UTP30 / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.668939 KDa |
| Sequence | String: MVESNDIIKS GLAEKALKAL ILQCEENPSL KNDKDIHIII NTGKKMGINR DNIPRIIPLT KYKLFKPRDL NILLITKDPS ALYRETLTK DEHTSELFKE IISVKNLRRR FKGSKLTQLY KDFDLVVADY RVHHLLPEVL GSRFYHGSKK LPYMIRMSKE V KLKRQQMV ...String: MVESNDIIKS GLAEKALKAL ILQCEENPSL KNDKDIHIII NTGKKMGINR DNIPRIIPLT KYKLFKPRDL NILLITKDPS ALYRETLTK DEHTSELFKE IISVKNLRRR FKGSKLTQLY KDFDLVVADY RVHHLLPEVL GSRFYHGSKK LPYMIRMSKE V KLKRQQMV EKCDPIYVRA QLRSICKNTS YIPNNDNCLS VRVGYIQKHS IPEILQNIQD TINFLTDKSK RPQGGVIKGG II SIFVKTS NSTSLPIYQF SEARENQKNE DLSDIKL |
+Macromolecule #28: Nucleolar protein 56
| Macromolecule | Name: Nucleolar protein 56 / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.961152 KDa |
| Sequence | String: MAPIEYLLFE EPTGYAVFKV KLQQDDIGSR LKEVQEQIND FGAFTKLIEL VSFAPFKGAA EALENANDIS EGLVSESLKA ILDLNLPKA SSKKKNITLA ISDKNLGPSI KEEFPYVDCI SNELAQDLIR GVRLHGEKLF KGLQSGDLER AQLGLGHAYS R AKVKFSVQ ...String: MAPIEYLLFE EPTGYAVFKV KLQQDDIGSR LKEVQEQIND FGAFTKLIEL VSFAPFKGAA EALENANDIS EGLVSESLKA ILDLNLPKA SSKKKNITLA ISDKNLGPSI KEEFPYVDCI SNELAQDLIR GVRLHGEKLF KGLQSGDLER AQLGLGHAYS R AKVKFSVQ KNDNHIIQAI ALLDQLDKDI NTFAMRVKEW YGWHFPELAK LVPDNYTFAK LVLFIKDKAS LNDDSLHDLA AL LNEDSGI AQRVIDNARI SMGQDISETD MENVCVFAQR VASLADYRRQ LYDYLCEKMH TVAPNLSELI GEVIGARLIS HAG SLTNLS KQAASTVQIL GAEKALFRAL KTKGNTPKYG LIYHSGFISK ASAKNKGRIS RYLANKCSMA SRIDNYSEEP SNVF GSVLK KQVEQRLEFY NTGKPTLKNE LAIQEAMELY NKDKPAAEVE ETKEKESSKK RKLEDDDEEK KEKKEKKSKK EKKEK KEKK DKKEKKDKKE KKDKKKKSKD |
+Macromolecule #29: Nucleolar protein 58
| Macromolecule | Name: Nucleolar protein 58 / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.060344 KDa |
| Sequence | String: MAYVLTETSA GYALLKASDK KIYKSSSLIQ DLDSSDKVLK EFKIAAFSKF NSAANALEEA NSIIEGKVSS QLEKLLEEIK KDKKSTLIV SETKLANAIN KLGLNFNVVS DAVTLDIYRA IKEYLPELLP GMSDNDLSKM SLGLAHSIGR HKLKFSADKV D VMIIQAIA ...String: MAYVLTETSA GYALLKASDK KIYKSSSLIQ DLDSSDKVLK EFKIAAFSKF NSAANALEEA NSIIEGKVSS QLEKLLEEIK KDKKSTLIV SETKLANAIN KLGLNFNVVS DAVTLDIYRA IKEYLPELLP GMSDNDLSKM SLGLAHSIGR HKLKFSADKV D VMIIQAIA LLDDLDKELN TYAMRCKEWY GWHFPELAKI VTDSVAYARI ILTMGIRSKA SETDLSEILP EEIEERVKTA AE VSMGTEI TQTDLDNINA LAEQIVEFAA YREQLSNYLS ARMKAIAPNL TQLVGELVGA RLIAHSGSLI SLAKSPASTI QIL GAEKAL FRALKTKHDT PKYGLLYHAS LVGQATGKNK GKIARVLAAK AAVSLRYDAL AEDRDDSGDI GLESRAKVEN RLSQ LEGRD LRTTPKVVRE AKKVEMTEAR AYNADADTAK AASDSESDSD DEEEEKKEKK EKKRKRDDDE DSKDSKKAKK EKKDK KEKK EKKEKKEKKE KKEKKEKKSK KEKKEKK |
+Macromolecule #30: 13 kDa ribonucleoprotein-associated protein
| Macromolecule | Name: 13 kDa ribonucleoprotein-associated protein / type: protein_or_peptide / ID: 30 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.582855 KDa |
| Sequence | String: MSAPNPKAFP LADAALTQQI LDVVQQAANL RQLKKGANEA TKTLNRGISE FIIMAADCEP IEILLHLPLL CEDKNVPYVF VPSRVALGR ACGVSRPVIA ASITTNDASA IKTQIYAVKD KIETLLI |
+Macromolecule #31: Ribosomal RNA-processing protein 9
| Macromolecule | Name: Ribosomal RNA-processing protein 9 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.146969 KDa |
| Sequence | String: MSDVTQQKKR KRSKGEVNPS KPTVDEEITD PSSNEDEQLE VSDEEDALES EEEFEGENPA DKRRRLAKQY LENLKSEAND ILTDNRNAE EKDLNNLKER TIDEYNNFDA GDLDKDIIAS RLKEDVAEQQ GRVFRYFGDK LLISEAKQSF TRVGENNLTC I SCFQPVLN ...String: MSDVTQQKKR KRSKGEVNPS KPTVDEEITD PSSNEDEQLE VSDEEDALES EEEFEGENPA DKRRRLAKQY LENLKSEAND ILTDNRNAE EKDLNNLKER TIDEYNNFDA GDLDKDIIAS RLKEDVAEQQ GRVFRYFGDK LLISEAKQSF TRVGENNLTC I SCFQPVLN KYTFEESSNG DKNKGRLFAY TVSKDLQLTK YDITDFSKRP KKLKYAKGGA KYIPTSKHEY ENTTEGHYDE IL TVAASPD GKYVVTGGRD RKLIVWSTES LSPVKVIPTK DRRGEVLSLA FRKNSDQLYA SCADFKIRTY SINQFSQLEI LYG HHDIVE DISALAMERC VTVGARDRTA MLWKIPDETR LTFRGGDEPQ KLLRRWMKEN AKEGEDGEVK YPDESEAPLF FCEG SIDVV SMVDDFHFIT GSDNGNICLW SLAKKKPIFT ERIAHGILPE PSFNDISGET DEELRKRQLQ GKKLLQPFWI TSLYA IPYS NVFISGSWSG SLKVWKISDN LRSFELLGEL SGAKGVVTKI QVVESGKHGK EKFRILASIA KEHRLGRWIA NVSGAR NGI YSAVIDQTGF |
+Macromolecule #32: U3 small nucleolar ribonucleoprotein protein IMP3
| Macromolecule | Name: U3 small nucleolar ribonucleoprotein protein IMP3 / type: protein_or_peptide / ID: 32 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.928529 KDa |
| Sequence | String: MVRKLKHHEQ KLLKKVDFLE WKQDQGHRDT QVMRTYHIQN REDYHKYNRI CGDIRRLANK LSLLPPTDPF RRKHEQLLLD KLYAMGVLT TKSKISDLEN KVTVSAICRR RLPVIMHRLK MAETIQDAVK FIEQGHVRVG PNLINDPAYL VTRNMEDYVT W VDNSKIKK TLLRYRNQID DFDFS |
+Macromolecule #33: U3 small nucleolar ribonucleoprotein protein IMP4
| Macromolecule | Name: U3 small nucleolar ribonucleoprotein protein IMP4 / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.536168 KDa |
| Sequence | String: MLRRQARERR EYLYRKAQEL QDSQLQQKRQ IIKQALAQGK PLPKELAEDE SLQKDFRYDQ SLKESEEADD LQVDDEYAAT SGIMDPRII VTTSRDPSTR LSQFAKEIKL LFPNAVRLNR GNYVMPNLVD ACKKSGTTDL VVLHEHRGVP TSLTISHFPH G PTAQFSLH ...String: MLRRQARERR EYLYRKAQEL QDSQLQQKRQ IIKQALAQGK PLPKELAEDE SLQKDFRYDQ SLKESEEADD LQVDDEYAAT SGIMDPRII VTTSRDPSTR LSQFAKEIKL LFPNAVRLNR GNYVMPNLVD ACKKSGTTDL VVLHEHRGVP TSLTISHFPH G PTAQFSLH NVVMRHDIIN AGNQSEVNPH LIFDNFTTAL GKRVVCILKH LFNAGPKKDS ERVITFANRG DFISVRQHVY VR TREGVEI AEVGPRFEMR LFELRLGTLE NKDADVEWQL RRFIRTANKK DYL |
+Macromolecule #34: U3 small nucleolar RNA-associated protein MPP10
| Macromolecule | Name: U3 small nucleolar RNA-associated protein MPP10 / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 67.042492 KDa |
| Sequence | String: MSELFGVLKS NAGRIILKDP SATSKDVKAY IDSVINTCKN GSITKKAELD EITVDGLDAN QVWWQVKLVL DSIDGDLIQG IQELKDVVT PSHNLSDGST LNSSSGEESE LEEAESVFKE KQMLSADVSE IEEQSNDSLS ENDEEPSMDD EKTSAEAARE E FAEEKRIS ...String: MSELFGVLKS NAGRIILKDP SATSKDVKAY IDSVINTCKN GSITKKAELD EITVDGLDAN QVWWQVKLVL DSIDGDLIQG IQELKDVVT PSHNLSDGST LNSSSGEESE LEEAESVFKE KQMLSADVSE IEEQSNDSLS ENDEEPSMDD EKTSAEAARE E FAEEKRIS SGQDERHSSP DPYGINDKFF DLEKFNRDTL AAEDSNEASE GSEDEDIDYF QDMPSDDEEE EAIYYEDFFD KP TKEPVKK HSDVKDPKED EELDEEEHDS AMDKVKLDLF ADEEDEPNAE GVGEASDKNL SSFEKQQIEI RKQIEQLENE AVA EKKWSL KGEVKAKDRP EDALLTEELE FDRTAKPVPV ITSEVTESLE DMIRRRIQDS NFDDLQRRTL LDITRKSQRP QFEL SDVKS SKSLAEIYED DYTRAEDESA LSEELQKAHS EISELYANLV YKLDVLSSVH FVPKPASTSL EIRVETPTIS MEDAQ PLYM SNASSLAPQE IYNVGKAEKD GEIRLKNGVA MSKEELTRED KNRLRRALKR KRSKANLPNV NKRSKRNDVV DTLSKA KNI TVINQKGEKK DVSGKTKKSR SGPDSTNIKL |
+Macromolecule #35: Ribosome biogenesis protein BMS1
| Macromolecule | Name: Ribosome biogenesis protein BMS1 / type: protein_or_peptide / ID: 35 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 135.792281 KDa |
| Sequence | String: MEQSNKQHRK AKEKNTAKKK LHTQGHNAKA FAVAAPGKMA RTMQRSSDVN ERKLHVPMVD RTPEDDPPPF IVAVVGPPGT GKTTLIRSL VRRMTKSTLN DIQGPITVVS GKHRRLTFLE CPADDLNAMI DIAKIADLVL LLIDGNFGFE METMEFLNIA Q HHGMPRVL ...String: MEQSNKQHRK AKEKNTAKKK LHTQGHNAKA FAVAAPGKMA RTMQRSSDVN ERKLHVPMVD RTPEDDPPPF IVAVVGPPGT GKTTLIRSL VRRMTKSTLN DIQGPITVVS GKHRRLTFLE CPADDLNAMI DIAKIADLVL LLIDGNFGFE METMEFLNIA Q HHGMPRVL GVATHLDLFK SQSTLRASKK RLKHRFWTEV YQGAKLFYLS GVINGRYPDR EILNLSRFIS VMKFRPLKWR NE HPYMLAD RFTDLTHPEL IETQGLQIDR KVAIYGYLHG TPLPSAPGTR VHIAGVGDFS VAQIEKLPDP CPTPFYQQKL DDF EREKMK EEAKANGEIT TASTTRRRKR LDDKDKLIYA PMSDVGGVLM DKDAVYIDIG KKNEEPSFVP GQERGEGEKL MTGL QSVEQ SIAEKFDGVG LQLFSNGTEL HEVADHEGMD VESGEESIED DEGKSKGRTS LRKPRIYGKP VQEEDADIDN LPSDE EPYT NDDDVQDSEP RMVEIDFNNT GEQGAEKLAL ETDSEFEESE DEFSWERTAA NKLKKTESKK RTWNIGKLIY MDNISP EEC IRRWRGEDDD SKDESDIEED VDDDFFRKKD GTVTKEGNKD HAVDLEKFVP YFDTFEKLAK KWKSVDAIKE RFLGAGI LG NDNKTKSDSN EGGEELYGDF EDLEDGNPSE QAEDNSDKES EDEDENEDTN GDDDNSFTNF DAEEKKDLTM EQEREMNA A KKEKLRAQFE IEEGENFKED DENNEYDTWY ELQKAKISKQ LEINNIEYQE MTPEQRQRIE GFKAGSYVRI VFEKVPMEF VKNFNPKFPI VMGGLLPTEI KFGIVKARLR RHRWHKKILK TNDPLVLSLG WRRFQTLPIY TTTDSRTRTR MLKYTPEHTY CNAAFYGPL CSPNTPFCGV QIVANSDTGN GFRIAATGIV EEIDVNIEIV KKLKLVGFPY KIFKNTAFIK DMFSSAMEVA R FEGAQIKT VSGIRGEIKR ALSKPEGHYR AAFEDKILMS DIVILRSWYP VRVKKFYNPV TSLLLKEKTE WKGLRLTGQI RA AMNLETP SNPDSAYHKI ERVERHFNGL KVPKAVQKEL PFKSQIHQMK PQKKKTYMAK RAVVLGGDEK KARSFIQKVL TIS KAKDSK RKEQKASQRK ERLKKLAKME EEKSQRDKEK KKEYFAQNGK RTTMGGDDES RPRKMRR |
+Macromolecule #36: RNA 3'-terminal phosphate cyclase-like protein
| Macromolecule | Name: RNA 3'-terminal phosphate cyclase-like protein / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.220559 KDa |
| Sequence | String: MSSSAPKYTT FQGSQNFRLR IVLATLSGKP IKIEKIRSGD LNPGLKDYEV SFLRLIESVT NGSVIEISYT GTTVIYRPGI IVGGASTHI CPSSKPVGYF VEPMLYLAPF SKKKFSILFK GITASHNDAG IEAIKWGLMP VMEKFGVREC ALHTLKRGSP P LGGGEVHL ...String: MSSSAPKYTT FQGSQNFRLR IVLATLSGKP IKIEKIRSGD LNPGLKDYEV SFLRLIESVT NGSVIEISYT GTTVIYRPGI IVGGASTHI CPSSKPVGYF VEPMLYLAPF SKKKFSILFK GITASHNDAG IEAIKWGLMP VMEKFGVREC ALHTLKRGSP P LGGGEVHL VVDSLIAQPI TMHEIDRPII SSITGVAYST RVSPSLVNRM IDGAKKVLKN LQCEVNITAD VWRGENSGKS PG WGITLVA QSKQKGWSYF AEDIGDAGSI PEELGEKVAC QLLEEISKSA AVGRNQLPLA IVYMVIGKED IGRLRINKEQ IDE RFIILL RDIKKIFNTE VFLKPVDEAD NEDMIATIKG IGFTNTSKKI A |
+Macromolecule #37: Ribosomal RNA-processing protein 7
| Macromolecule | Name: Ribosomal RNA-processing protein 7 / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.526441 KDa |
| Sequence | String: MGIEDISAMK NGFIVVPFKL PDHKALPKSQ EASLHFMFAK RHQSSNSNES DCLFLVNLPL LSNIEHMKKF VGQLCGKYDT VSHVEELLY NDEFGLHEVD LSALTSDLMS STDVNEKRYT PRNTALLKFV DAASINNCWN ALKKYSNLHA KHPNELFEWT Y TTPSFTTF ...String: MGIEDISAMK NGFIVVPFKL PDHKALPKSQ EASLHFMFAK RHQSSNSNES DCLFLVNLPL LSNIEHMKKF VGQLCGKYDT VSHVEELLY NDEFGLHEVD LSALTSDLMS STDVNEKRYT PRNTALLKFV DAASINNCWN ALKKYSNLHA KHPNELFEWT Y TTPSFTTF VNFYKPLDID YLKEDIHTHM AIFEQREAQA QEDVQSSIVD EDGFTLVVGK NTKSLNSIRK KILNKNPLSK HE NKAKPIS NIDKKAKKDF YRFQVRERKK QEINQLLSKF KEDQERIKVM KAKRKFNPYT |
+Macromolecule #38: Ribosome biogenesis protein ENP2
| Macromolecule | Name: Ribosome biogenesis protein ENP2 / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.864172 KDa |
| Sequence | String: MVLKSTSAND VSVYQVSGTN VSRSLPDWIA KKRKRQLKND LEYQNRVELI QDFEFSEASN KIKVSRDGQY CMATGTYKPQ IHVYDFANL SLKFDRHTDA ENVDFTILSD DWTKSVHLQN DRSIQFQNKG GLHYTTRIPK FGRSLVYNKV NCDLYVGASG N ELYRLNLE ...String: MVLKSTSAND VSVYQVSGTN VSRSLPDWIA KKRKRQLKND LEYQNRVELI QDFEFSEASN KIKVSRDGQY CMATGTYKPQ IHVYDFANL SLKFDRHTDA ENVDFTILSD DWTKSVHLQN DRSIQFQNKG GLHYTTRIPK FGRSLVYNKV NCDLYVGASG N ELYRLNLE KGRFLNPFKL DTEGVNHVSI NEVNGLLAAG TETNVVEFWD PRSRSRVSKL YLENNIDNRP FQVTTTSFRN DG LTFACGT SNGYSYIYDL RTSEPSIIKD QGYGFDIKKI IWLDNVGTEN KIVTCDKRIA KIWDRLDGKA YASMEPSVDI NDI EHVPGT GMFFTANESI PMHTYYIPSL GPSPRWCSFL DSITEELEEK PSDTVYSNYR FITRDDVKKL NLTHLVGSRV LRAY MHGFF INTELYDKVS LIANPDAYKD EREREIRRRI EKERESRIRS SGAVQKPKIK VNKTLVDKLS QKRGDKVAGK VLTDD RFKE MFEDEEFQVD EDDYDFKQLN PVKSIKETEE GAAKRIRALT AAEESDEERI AMKDGRGHYD YEDEESDEEE SDDETN QKS NKEELSEKDL RKMEKQKALI ERRKKEKEQS ERFMNEMKAG TSTSTQRDES AHVTFGEQVG ELLEVENGKK SNESILR RN QRGEAELTFI PQRKSKKDGN YKSRRHDNSS DEEGIDENGN KKDNGRSKPR FENRRRASKN AFRGM |
+Macromolecule #39: Ribosomal RNA small subunit methyltransferase NEP1
| Macromolecule | Name: Ribosomal RNA small subunit methyltransferase NEP1 / type: protein_or_peptide / ID: 39 / Number of copies: 2 / Enantiomer: LEVO EC number: rRNA small subunit pseudouridine methyltransferase Nep1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.936461 KDa |
| Sequence | String: MVEDSRVRDA LKGGDQKALP ASLVPQAPPV LTSKDKITKR MIVVLAMASL ETHKISSNGP GGDKYVLLNC DDHQGLLKKM GRDISEARP DITHQCLLTL LDSPINKAGK LQVYIQTSRG ILIEVNPTVR IPRTFKRFSG LMVQLLHKLS IRSVNSEEKL L KVIKNPIT ...String: MVEDSRVRDA LKGGDQKALP ASLVPQAPPV LTSKDKITKR MIVVLAMASL ETHKISSNGP GGDKYVLLNC DDHQGLLKKM GRDISEARP DITHQCLLTL LDSPINKAGK LQVYIQTSRG ILIEVNPTVR IPRTFKRFSG LMVQLLHKLS IRSVNSEEKL L KVIKNPIT DHLPTKCRKV TLSFDAPVIR VQDYIEKLDD DESICVFVGA MARGKDNFAD EYVDEKVGLS NYPLSASVAC SK FCHGAED AWNIL |
+Macromolecule #40: Essential nuclear protein 1
| Macromolecule | Name: Essential nuclear protein 1 / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.207422 KDa |
| Sequence | String: MARASSTKAR KQRHDPLLKD LDAAQGTLKK INKKKLAQND AANHDAANEE DGYIDSKASR KILQLAKEQQ DEIEGEELAE SERNKQFEA RFTTMSYDDE DEDEDEDEEA FGEDISDFEP EGDYKEEEEI VEIDEEDAAM FEQYFKKSDD FNSLSGSYNL A DKIMASIR ...String: MARASSTKAR KQRHDPLLKD LDAAQGTLKK INKKKLAQND AANHDAANEE DGYIDSKASR KILQLAKEQQ DEIEGEELAE SERNKQFEA RFTTMSYDDE DEDEDEDEEA FGEDISDFEP EGDYKEEEEI VEIDEEDAAM FEQYFKKSDD FNSLSGSYNL A DKIMASIR EKESQVEDMQ DDEPLANEQN TSRGNISSGL KSGEGVALPE KVIKAYTTVG SILKTWTHGK LPKLFKVIPS LR NWQDVIY VTNPEEWSPH VVYEATKLFV SNLTAKESQK FINLILLERF RDNIETSEDH SLNYHIYRAV KKSLYKPSAF FKG FLFPLV ETGCNVREAT IAGSVLAKVS VPALHSSAAL SYLLRLPFSP PTTVFIKILL DKKYALPYQT VDDCVYYFMR FRIL DDGSN GEDATRVLPV IWHKAFLTFA QRYKNDITQD QRDFLLETVR QRGHKDIGPE IRRELLAGAS REFVDPQEAN DDLMI DVN |
+Macromolecule #41: rRNA biogenesis protein RRP5
| Macromolecule | Name: rRNA biogenesis protein RRP5 / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 193.411422 KDa |
| Sequence | String: MVASTKRKRD EDFPLSREDS TKQPSTSSLV RNTEEVSFPR GGASALTPLE LKQVANEAAS DVLFGNESVK ASEPASRPLK KKKTTKKST SKDSEASSAN SDEARAGLIE HVNFKTLKNG SSLLGQISAI TKQDLCITFT DGISGYVNLT HISEEFTSIL E DLDEDMDS ...String: MVASTKRKRD EDFPLSREDS TKQPSTSSLV RNTEEVSFPR GGASALTPLE LKQVANEAAS DVLFGNESVK ASEPASRPLK KKKTTKKST SKDSEASSAN SDEARAGLIE HVNFKTLKNG SSLLGQISAI TKQDLCITFT DGISGYVNLT HISEEFTSIL E DLDEDMDS DTDAADEKKS KVEDAEYESS DDEDEKLDKS NELPNLRRYF HIGQWLRCSV IKNTSLEPST KKSKKKRIEL TI EPSSVNI YADEDLVKST SIQCAVKSIE DHGATLDVGL PGFTGFIAKK DFGNFEKLLP GAVFLGNITK KSDRSIVVNT DFS DKKNKI TQISSIDAII PGQIVDLLCE SITKNGIAGK VFGLVSGVVN VSHLRTFSEE DLKHKFVIGS SIRCRIIACL ENKS GDKVL ILSNLPHILK LEDALRSTEG LDAFPIGYTF ESCSIKGRDS EYLYLALDDD RLGKVHSSRV GEIENSENLS SRVLG YSPV DDIYQLSTDP KYLKLKYLRT NDIPIGELLP SCEITSVSSS GIELKIFNGQ FKASVPPLHI SDTRLVYPER KFKIGS KVK GRVISVNSRG NVHVTLKKSL VNIEDNELPL VSTYENAKNI KEKNEKTLAT IQVFKPNGCI ISFFGGLSGF LPNSEIS EV FVKRPEEHLR LGQTVIVKLL DVDADRRRII ATCKVSNEQA AQQKDTIENI VPGRTIITVH VIEKTKDSVI VEIPDVGL R GVIYVGHLSD SRIEQNRAQL KKLRIGTELT GLVIDKDTRT RVFNMSLKSS LIKDAKKETL PLTYDDVKDL NKDVPMHAY IKSISDKGLF VAFNGKFIGL VLPSYAVDSR DIDISKAFYI NQSVTVYLLR TDDKNQKFLL SLKAPKVKEE KKKVESNIED PVDSSIKSW DDLSIGSIVK AKIKSVKKNQ LNVILAANLH GRVDIAEVFD TYEEITDKKQ PLSNYKKDDV IKVKIIGNHD V KSHKFLPI THKISKASVL ELSMKPSELK SKEVHTKSLE EINIGQELTG FVNNSSGNHL WLTISPVLKA RISLLDLADN DS NFSENIE SVFPLGSALQ VKVASIDREH GFVNAIGKSH VDINMSTIKV GDELPGRVLK IAEKYVLLDL GNKVTGISFI TDA LNDFSL TLKEAFEDKI NNVIPTTVLS VDEQNKKIEL SLRPATAKTR SIKSHEDLKQ GEIVDGIVKN VNDKGIFVYL SRKV EAFVP VSKLSDSYLK EWKKFYKPMQ YVLGKVVTCD EDSRISLTLR ESEINGDLKV LKTYSDIKAG DVFEGTIKSV TDFGV FVKL DNTVNVTGLA HITEIADKKP EDLSALFGVG DRVKAIVLKT NPEKKQISLS LKASHFSKEA ELASTTTTTT TVDQLE KED EDEVMADAGF NDSDSESDIG DQNTEVADRK PETSSDGLSL SAGFDWTASI LDQAQEEEES DQDQEDFTEN KKHKHKR RK ENVVQDKTID INTRAPESVA DFERLLIGNP NSSVVWMNYM AFQLQLSEIE KARELAERAL KTINFREEAE KLNIWIAM L NLENTFGTEE TLEEVFSRAC QYMDSYTIHT KLLGIYEISE KFDKAAELFK ATAKKFGGEK VSIWVSWGDF LISHNEEQE ARTILGNALK ALPKRNHIEV VRKFAQLEFA KGDPERGRSL FEGLVADAPK RIDLWNVYVD QEVKAKDKKK VEDLFERIIT KKITRKQAK FFFNKWLQFE ESEGDEKTIE YVKAKATEYV ASHESQKADE |
+Macromolecule #42: Pre-rRNA-processing protein PNO1
| Macromolecule | Name: Pre-rRNA-processing protein PNO1 / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.380623 KDa |
| Sequence | String: MVAPTALKKA TVTPVSGQDG GSSRIIGINN TESIDEDDDD DVLLDDSDNN TAKEEVEGEE GSRKTHESKT VVVDDQGKPR FTSASKTQG NKIKFESRKI MVPPHRMTPL RNSWTKIYPP LVEHLKLQVR MNLKTKSVEL RTNPKFTTDP GALQKGADFI K AFTLGFDL ...String: MVAPTALKKA TVTPVSGQDG GSSRIIGINN TESIDEDDDD DVLLDDSDNN TAKEEVEGEE GSRKTHESKT VVVDDQGKPR FTSASKTQG NKIKFESRKI MVPPHRMTPL RNSWTKIYPP LVEHLKLQVR MNLKTKSVEL RTNPKFTTDP GALQKGADFI K AFTLGFDL DDSIALLRLD DLYIETFEVK DVKTLTGDHL SRAIGRIAGK DGKTKFAIEN ATRTRIVLAD SKIHILGGFT HI RMARESV VSLILGSPPG KVYGNLRTVA SRLKERY |
+Macromolecule #43: Protein BFR2
| Macromolecule | Name: Protein BFR2 / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.285039 KDa |
| Sequence | String: MEKSLADQIS DIAIKPVNKD FDIEDEENAS LFQHNEKNGE SDLSDYGNSN TEETKKAHYL EVEKSKLRAE KGLELNDPKY TGVKGSRQA LYEEVSENED EEEEEEEEEE KEEDALSFRT DSEDEEVEID EEESDADGGE TEEAQQKRHA LSKLIQQETK Q AINKLSQS ...String: MEKSLADQIS DIAIKPVNKD FDIEDEENAS LFQHNEKNGE SDLSDYGNSN TEETKKAHYL EVEKSKLRAE KGLELNDPKY TGVKGSRQA LYEEVSENED EEEEEEEEEE KEEDALSFRT DSEDEEVEID EEESDADGGE TEEAQQKRHA LSKLIQQETK Q AINKLSQS VQRDASKGYS ILQQTKLFDN IIDLRIKLQK AVIAANKLPL TTESWEEAKM DDSEETKRLL KENEKLFNNL FN RLINFRI KFQLGDHITQ NEEVAKHKLS KKRSLKELYQ ETNSLDSELK EYRTAVLNKW STKVSSASGN AALSSNKFKA INL PADVQV ENQLSDMSRL MKRTKLNRRN ITPLYFQKDC ANGRLPELIS PVVKDSVDDN ENSDDGLDIP KNYDPRRKDN NAID ITENP YVFDDEDFYR VLLNDLIDKK ISNAHNSESA AITITSTNAR SNNKLKKNID TKASKGRKLN YSVQDPIANY EAPIT SGYK WSDDQIDEFF AGLLGQRVNF NENEDEEQHA RIENDEELEA VKNDDIQIFG |
+Macromolecule #44: rRNA-processing protein FCF2
| Macromolecule | Name: rRNA-processing protein FCF2 / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.68924 KDa |
| Sequence | String: MDQSVEDLFG ALRDASASLE VKNSAKEQVS LQQEDVLQIG NNDDEVEIES KFQEIETNLK KLPKLETGFD ALANKKKKKN VLPSVETED KRKPNKSDKN DNDWFTLPKP DDNMRREVQR DLLLIKHRAA LDPKRHYKKQ RWEVPERFAI GTIIEDKSEF Y SSRMNRKE ...String: MDQSVEDLFG ALRDASASLE VKNSAKEQVS LQQEDVLQIG NNDDEVEIES KFQEIETNLK KLPKLETGFD ALANKKKKKN VLPSVETED KRKPNKSDKN DNDWFTLPKP DDNMRREVQR DLLLIKHRAA LDPKRHYKKQ RWEVPERFAI GTIIEDKSEF Y SSRMNRKE RKSTILETLM GDEASNKYFK RKYNEIQEKS TSGRKAHYKK MKEMRKKRR |
+Macromolecule #45: Protein FAF1
| Macromolecule | Name: Protein FAF1 / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.961512 KDa |
| Sequence | String: MTLDDDDYIK QMELQRKAFE SQFGSLESMG FEDKTKNIRT EVDTRDSSGD EIDNSDHGSD FKDGTIESSN SSDEDSGNET AEENNQDSK PKTQPKVIRF NGPSDVYVPP SKKTQKLLRS GKTLTQINKK LESTEAKEEK EDETLEAENL QNDLELQQFL R ESHLLSAF ...String: MTLDDDDYIK QMELQRKAFE SQFGSLESMG FEDKTKNIRT EVDTRDSSGD EIDNSDHGSD FKDGTIESSN SSDEDSGNET AEENNQDSK PKTQPKVIRF NGPSDVYVPP SKKTQKLLRS GKTLTQINKK LESTEAKEEK EDETLEAENL QNDLELQQFL R ESHLLSAF NNGGSGSTNS GVSLTLQSMG GGNDDGIVYQ DDQVIGKARS RTLEMRLNRL SRVNGHQDKI NKLEKVPMHI RR GMIDKHV KRIKKYEQEA AEGGIVLSKV KKGQFRKIES TYKKDIERRI GGSIKARDKE KATKRERGLK ISSVGRSTRN GLI VSKRDI ARISGGERSG KFNGKKKSRR |
+Macromolecule #46: KRR1 small subunit processome component
| Macromolecule | Name: KRR1 small subunit processome component / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.226254 KDa |
| Sequence | String: MVSTHNRDKP WDTDDIDKWK IEEFKEEDNA SGQPFAEESS FMTLFPKYRE SYLKTIWNDV TRALDKHNIA CVLDLVEGSM TVKTTRKTY DPAIILKARD LIKLLARSVP FPQAVKILQD DMACDVIKIG NFVTNKERFV KRRQRLVGPN GNTLKALELL T KCYILVQG ...String: MVSTHNRDKP WDTDDIDKWK IEEFKEEDNA SGQPFAEESS FMTLFPKYRE SYLKTIWNDV TRALDKHNIA CVLDLVEGSM TVKTTRKTY DPAIILKARD LIKLLARSVP FPQAVKILQD DMACDVIKIG NFVTNKERFV KRRQRLVGPN GNTLKALELL T KCYILVQG NTVSAMGPFK GLKEVRRVVE DCMKNIHPIY HIKELMIKRE LAKRPELANE DWSRFLPMFK KRNVARKKPK KI RNVEKKV YTPFPPAQLP RKVDLEIESG EYFLSKREKQ MKKLNEQKEK QMEREIERQE ERAKDFIAPE EEAYKPNQN |
+Macromolecule #47: Protein SOF1
| Macromolecule | Name: Protein SOF1 / type: protein_or_peptide / ID: 47 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.888918 KDa |
| Sequence | String: MKIKTIKRSA DDYVPVKSTQ ESQMPRNLNP ELHPFERARE YTKALNATKL ERMFAKPFVG QLGYGHRDGV YAIAKNYGSL NKLATGSAD GVIKYWNMST REEFVSFKAH YGLVTGLCVT QPRFHDKKPD LKSQNFMLSC SDDKTVKLWS INVDDYSNKN S SDNDSVTN ...String: MKIKTIKRSA DDYVPVKSTQ ESQMPRNLNP ELHPFERARE YTKALNATKL ERMFAKPFVG QLGYGHRDGV YAIAKNYGSL NKLATGSAD GVIKYWNMST REEFVSFKAH YGLVTGLCVT QPRFHDKKPD LKSQNFMLSC SDDKTVKLWS INVDDYSNKN S SDNDSVTN EEGLIRTFDG ESAFQGIDSH RENSTFATGG AKIHLWDVNR LKPVSDLSWG ADNITSLKFN QNETDILAST GS DNSIVLY DLRTNSPTQK IVQTMRTNAI CWNPMEAFNF VTANEDHNAY YYDMRNLSRS LNVFKDHVSA VMDVDFSPTG DEI VTGSYD KSIRIYKTNH GHSREIYHTK RMQHVFQVKY SMDSKYIISG SDDGNVRLWR SKAWERSNVK TTREKNKLEY DEKL KERFR HMPEIKRISR HRHVPQVIKK AQEIKNIELS SIKRREANER RTRKDMPYIS ERKKQIVGTV HKYEDSGRDR KRRKE DDKR DTQEK |
+Macromolecule #48: Regulator of rDNA transcription protein 14
| Macromolecule | Name: Regulator of rDNA transcription protein 14 / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.898289 KDa |
| Sequence | String: MSSSLSQTSK YQATSVVNGL LSNLLPGVPK IRANNGKTSV NNGSKAQLID RNLKKRVQLQ NRDVHKIKKK CKLVKKKKVK KHKLDKEQL EQLAKHQVLK KHQHEGTLTD HERKYLNKLI KRNSQNLRSW DLEEEVRDEL EDIQQSILKD TVSTANTDRS K RRRFKRKQ ...String: MSSSLSQTSK YQATSVVNGL LSNLLPGVPK IRANNGKTSV NNGSKAQLID RNLKKRVQLQ NRDVHKIKKK CKLVKKKKVK KHKLDKEQL EQLAKHQVLK KHQHEGTLTD HERKYLNKLI KRNSQNLRSW DLEEEVRDEL EDIQQSILKD TVSTANTDRS K RRRFKRKQ FKEDIKESDF VKDHRYPGLT PGLAPVGLSD EEDSSEED |
+Macromolecule #49: 40S ribosomal protein S4-A
| Macromolecule | Name: 40S ribosomal protein S4-A / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.46933 KDa |
| Sequence | String: MARGPKKHLK RLAAPHHWLL DKLSGCYAPR PSAGPHKLRE SLPLIVFLRN RLKYALNGRE VKAILMQRHV KVDGKVRTDT TYPAGFMDV ITLDATNENF RLVYDVKGRF AVHRITDEEA SYKLGKVKKV QLGKKGVPYV VTHDGRTIRY PDPNIKVNDT V KIDLASGK ...String: MARGPKKHLK RLAAPHHWLL DKLSGCYAPR PSAGPHKLRE SLPLIVFLRN RLKYALNGRE VKAILMQRHV KVDGKVRTDT TYPAGFMDV ITLDATNENF RLVYDVKGRF AVHRITDEEA SYKLGKVKKV QLGKKGVPYV VTHDGRTIRY PDPNIKVNDT V KIDLASGK ITDFIKFDAG KLVYVTGGRN LGRIGTIVHK ERHDGGFDLV HIKDSLDNTF VTRLNNVFVI GEQGKPYISL PK GKGIKLS IAEERDRRRA QQGL |
+Macromolecule #50: 40S ribosomal protein S5
| Macromolecule | Name: 40S ribosomal protein S5 / type: protein_or_peptide / ID: 50 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.0726 KDa |
| Sequence | String: MSDTEAPVEV QEDFEVVEEF TPVVLATPIP EEVQQAQTEI KLFNKWSFEE VEVKDASLVD YVQVRQPIFV AHTAGRYANK RFRKAQCPI IERLTNSLMM NGRNNGKKLK AVRIIKHTLD IINVLTDQNP IQVVVDAITN TGPREDTTRV GGGGAARRQA V DVSPLRRV ...String: MSDTEAPVEV QEDFEVVEEF TPVVLATPIP EEVQQAQTEI KLFNKWSFEE VEVKDASLVD YVQVRQPIFV AHTAGRYANK RFRKAQCPI IERLTNSLMM NGRNNGKKLK AVRIIKHTLD IINVLTDQNP IQVVVDAITN TGPREDTTRV GGGGAARRQA V DVSPLRRV NQAIALLTIG AREAAFRNIK TIAETLAEEL INAAKGSSTS YAIKKKDELE RVAKSNR |
+Macromolecule #51: 40S ribosomal protein S6-A
| Macromolecule | Name: 40S ribosomal protein S6-A / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.054486 KDa |
| Sequence | String: MKLNISYPVN GSQKTFEIDD EHRIRVFFDK RIGQEVDGEA VGDEFKGYVF KISGGNDKQG FPMKQGVLLP TRIKLLLTKN VSCYRPRRD GERKRKSVRG AIVGPDLAVL ALVIVKKGEQ ELEGLTDTTV PKRLGPKRAN NIRKFFGLSK EDDVRDFVIR R EVTKGEKT ...String: MKLNISYPVN GSQKTFEIDD EHRIRVFFDK RIGQEVDGEA VGDEFKGYVF KISGGNDKQG FPMKQGVLLP TRIKLLLTKN VSCYRPRRD GERKRKSVRG AIVGPDLAVL ALVIVKKGEQ ELEGLTDTTV PKRLGPKRAN NIRKFFGLSK EDDVRDFVIR R EVTKGEKT YTKAPKIQRL VTPQRLQRKR HQRALKVRNA QAQREAAAEY AQLLAKRLSE RKAEKAEIRK RRASSLKA |
+Macromolecule #52: 40S ribosomal protein S7-A
| Macromolecule | Name: 40S ribosomal protein S7-A / type: protein_or_peptide / ID: 52 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.658209 KDa |
| Sequence | String: MSAPQAKILS QAPTELELQV AQAFVELENS SPELKAELRP LQFKSIREID VAGGKKALAI FVPVPSLAGF HKVQTKLTRE LEKKFQDRH VIFLAERRIL PKPSRTSRQV QKRPRSRTLT AVHDKILEDL VFPTEIVGKR VRYLVGGNKI QKVLLDSKDV Q QIDYKLES FQAVYNKLTG KQIVFEIPSE TH |
+Macromolecule #53: 40S ribosomal protein S8-A
| Macromolecule | Name: 40S ribosomal protein S8-A / type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.537803 KDa |
| Sequence | String: MGISRDSRHK RSATGAKRAQ FRKKRKFELG RQPANTKIGA KRIHSVRTRG GNKKYRALRI ETGNFSWASE GISKKTRIAG VVYHPSNNE LVRTNTLTKA AIVQIDATPF RQWFEAHYGQ TLGKKKNVKE EETVAKSKNA ERKWAARAAS AKIESSVESQ F SAGRLYAC ...String: MGISRDSRHK RSATGAKRAQ FRKKRKFELG RQPANTKIGA KRIHSVRTRG GNKKYRALRI ETGNFSWASE GISKKTRIAG VVYHPSNNE LVRTNTLTKA AIVQIDATPF RQWFEAHYGQ TLGKKKNVKE EETVAKSKNA ERKWAARAAS AKIESSVESQ F SAGRLYAC ISSRPGQSGR CDGYILEGEE LAFYLRRLTA KK |
+Macromolecule #54: 40S ribosomal protein S9-A
| Macromolecule | Name: 40S ribosomal protein S9-A / type: protein_or_peptide / ID: 54 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.487893 KDa |
| Sequence | String: MPRAPRTYSK TYSTPKRPYE SSRLDAELKL AGEFGLKNKK EIYRISFQLS KIRRAARDLL TRDEKDPKRL FEGNALIRRL VRVGVLSED KKKLDYVLAL KVEDFLERRL QTQVYKLGLA KSVHHARVLI TQRHIAVGKQ IVNIPSFMVR LDSEKHIDFA P TSPFGGAR ...String: MPRAPRTYSK TYSTPKRPYE SSRLDAELKL AGEFGLKNKK EIYRISFQLS KIRRAARDLL TRDEKDPKRL FEGNALIRRL VRVGVLSED KKKLDYVLAL KVEDFLERRL QTQVYKLGLA KSVHHARVLI TQRHIAVGKQ IVNIPSFMVR LDSEKHIDFA P TSPFGGAR PGRVARRNAA RKAEASGEAA DEADEADEE |
+Macromolecule #55: 40S ribosomal protein S11-A
| Macromolecule | Name: 40S ribosomal protein S11-A / type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.785934 KDa |
| Sequence | String: MSTELTVQSE RAFQKQPHIF NNPKVKTSKR TKRWYKNAGL GFKTPKTAIE GSYIDKKCPF TGLVSIRGKI LTGTVVSTKM HRTIVIRRA YLHYIPKYNR YEKRHKNVPV HVSPAFRVQV GDIVTVGQCR PISKTVRFNV VKVSAAAGKA NKQFAKF |
+Macromolecule #56: 40S ribosomal protein S13
| Macromolecule | Name: 40S ribosomal protein S13 / type: protein_or_peptide / ID: 56 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.059945 KDa |
| Sequence | String: MGRMHSAGKG ISSSAIPYSR NAPAWFKLSS ESVIEQIVKY ARKGLTPSQI GVLLRDAHGV TQARVITGNK IMRILKSNGL APEIPEDLY YLIKKAVSVR KHLERNRKDK DAKFRLILIE SRIHRLARYY RTVAVLPPNW KYESATASAL VN |
+Macromolecule #57: 40S ribosomal protein S14-A
| Macromolecule | Name: 40S ribosomal protein S14-A / type: protein_or_peptide / ID: 57 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.562655 KDa |
| Sequence | String: MSNVVQARDN SQVFGVARIY ASFNDTFVHV TDLSGKETIA RVTGGMKVKA DRDESSPYAA MLAAQDVAAK CKEVGITAVH VKIRATGGT RTKTPGPGGQ AALRALARSG LRIGRIEDVT PVPSDSTRKK GGRRGRRL |
+Macromolecule #58: 40S ribosomal protein S16-A
| Macromolecule | Name: 40S ribosomal protein S16-A / type: protein_or_peptide / ID: 58 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.87749 KDa |
| Sequence | String: MSAVPSVQTF GKKKSATAVA HVKAGKGLIK VNGSPITLVE PEILRFKVYE PLLLVGLDKF SNIDIRVRVT GGGHVSQVYA IRQAIAKGL VAYHQKYVDE QSKNELKKAF TSYDRTLLIA DSRRPEPKKF GGKGARSRFQ KSYR |
+Macromolecule #59: 40S ribosomal protein S18-A
| Macromolecule | Name: 40S ribosomal protein S18-A / type: protein_or_peptide / ID: 59 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.071641 KDa |
| Sequence | String: MSLVVQEQGS FQHILRLLNT NVDGNIKIVY ALTTIKGVGR RYSNLVCKKA DVDLHKRAGE LTQEELERIV QIMQNPTHYK IPAWFLNRQ NDITDGKDYH TLANNVESKL RDDLERLKKI RAHRGIRHFW GLRVRGQHTK TTGRRRA |
+Macromolecule #60: 40S ribosomal protein S22-A
| Macromolecule | Name: 40S ribosomal protein S22-A / type: protein_or_peptide / ID: 60 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.650062 KDa |
| Sequence | String: MTRSSVLADA LNAINNAEKT GKRQVLIRPS SKVIIKFLQV MQKHGYIGEF EYIDDHRSGK IVVQLNGRLN KCGVISPRFN VKIGDIEKW TANLLPARQF GYVILTTSAG IMDHEEARRK HVSGKILGFV Y |
+Macromolecule #61: 40S ribosomal protein S23-A
| Macromolecule | Name: 40S ribosomal protein S23-A / type: protein_or_peptide / ID: 61 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.073896 KDa |
| Sequence | String: MGKGKPRGLN SARKLRVHRR NNRWAENNYK KRLLGTAFKS SPFGGSSHAK GIVLEKLGIE SKQPNSAIRK CVRVQLIKNG KKVTAFVPN DGCLNFVDEN DEVLLAGFGR KGKAKGDIPG VRFKVVKVSG VSLLALWKEK KEKPRS |
+Macromolecule #62: 40S ribosomal protein S24-A
| Macromolecule | Name: 40S ribosomal protein S24-A / type: protein_or_peptide / ID: 62 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.362848 KDa |
| Sequence | String: MSDAVTIRTR KVISNPLLAR KQFVVDVLHP NRANVSKDEL REKLAEVYKA EKDAVSVFGF RTQFGGGKSV GFGLVYNSVA EAKKFEPTY RLVRYGLAEK VEKASRQQRK QKKNRDKKIF GTGKRLAKKV ARRNAD |
+Macromolecule #63: 40S ribosomal protein S27-A
| Macromolecule | Name: 40S ribosomal protein S27-A / type: protein_or_peptide / ID: 63 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.893391 KDa |
| Sequence | String: MVLVQDLLHP TAASEARKHK LKTLVQGPRS YFLDVKCPGC LNITTVFSHA QTAVTCESCS TILCTPTGGK AKLSEGTSFR RK |
+Macromolecule #64: 40S ribosomal protein S28-A
| Macromolecule | Name: 40S ribosomal protein S28-A / type: protein_or_peptide / ID: 64 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.605847 KDa |
| Sequence | String: MDNKTPVTLA KVIKVLGRTG SRGGVTQVRV EFLEDTSRTI VRNVKGPVRE NDILVLMESE REARRLR |
+Macromolecule #69: Exosome complex component RRP45
| Macromolecule | Name: Exosome complex component RRP45 / type: protein_or_peptide / ID: 69 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.001859 KDa |
| Sequence | String: MAKDIEISAS ESKFILEALR QNYRLDGRSF DQFRDVEITF GKEFGDVSVK MGNTKVHCRI SCQIAQPYED RPFEGLFVIS TEISPMAGS QFENGNITGE DEVLCSRIIE KSVRRSGALD VEGLCIVAGS KCWAVRADVH FLDCDGGFID ASCIAVMAGL M HFKKPDIT ...String: MAKDIEISAS ESKFILEALR QNYRLDGRSF DQFRDVEITF GKEFGDVSVK MGNTKVHCRI SCQIAQPYED RPFEGLFVIS TEISPMAGS QFENGNITGE DEVLCSRIIE KSVRRSGALD VEGLCIVAGS KCWAVRADVH FLDCDGGFID ASCIAVMAGL M HFKKPDIT VHGEQIIVHP VNEREPVPLG ILHIPICVTF SFFNPQDTEE NIKGETNSEI SIIDATLKEE LLRDGVLTVT LN KNREVVQ VSKAGGLPMD ALTLMKCCHE AYSIIEKITD QILQLLKEDS EKRNKYAAML TSENAREI |
+Macromolecule #70: Exosome complex component SKI6
| Macromolecule | Name: Exosome complex component SKI6 / type: protein_or_peptide / ID: 70 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.599727 KDa |
| Sequence | String: MSRLEIYSPE GLRLDGRRWN ELRRFESSIN THPHAADGSS YMEQGNNKII TLVKGPKEPR LKSQMDTSKA LLNVSVNITK FSKFERSKS SHKNERRVLE IQTSLVRMFE KNVMLNIYPR TVIDIEIHVL EQDGGIMGSL INGITLALID AGISMFDYIS G ISVGLYDT ...String: MSRLEIYSPE GLRLDGRRWN ELRRFESSIN THPHAADGSS YMEQGNNKII TLVKGPKEPR LKSQMDTSKA LLNVSVNITK FSKFERSKS SHKNERRVLE IQTSLVRMFE KNVMLNIYPR TVIDIEIHVL EQDGGIMGSL INGITLALID AGISMFDYIS G ISVGLYDT TPLLDTNSLE ENAMSTVTLG VVGKSEKLSL LLVEDKIPLD RLENVLAIGI AGAHRVRDLM DEELRKHAQK RV SNASAR |
+Macromolecule #71: Exosome complex component RRP43
| Macromolecule | Name: Exosome complex component RRP43 / type: protein_or_peptide / ID: 71 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.093 KDa |
| Sequence | String: MAESTTLETI EIHPITFPPE VLARISPELS LQRHLSLGIR PCLRKYEEFR DVAIENNTLS RYADAGNIDT KNNILGSNVL KSGKTIVIT SITGGIIEET SAAIKDLDDF GEEELFEVTK EEDIIANYAS VYPVVEVERG RVGACTDEEM TISQKLHDSI L HSRILPKK ...String: MAESTTLETI EIHPITFPPE VLARISPELS LQRHLSLGIR PCLRKYEEFR DVAIENNTLS RYADAGNIDT KNNILGSNVL KSGKTIVIT SITGGIIEET SAAIKDLDDF GEEELFEVTK EEDIIANYAS VYPVVEVERG RVGACTDEEM TISQKLHDSI L HSRILPKK ALKVKAGVRS ANEDGTFSVL YPDELEDDTL NETNLKMKRK WSYVLYAKIV VLSRTGPVFD LCWNSLMYAL QS VKLPRAF IDERASDLRM TIRTRGRSAT IRETYEIICD QTKSVPLMIN AKNIAFASNY GIVELDPECQ LQNSDNSEEE EVD IDMDKL NTVLIADLDT EAEETSIHST ISILAAPSGN YKQLTLMGGG AKITPEMIKR SLLLSRVRAD DLSTRFNI |
+Macromolecule #72: Exosome complex component RRP46
| Macromolecule | Name: Exosome complex component RRP46 / type: protein_or_peptide / ID: 72 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.430193 KDa |
| Sequence | String: MSVQAEIGIL DHVDGSSEFV SQDTKVICSV TGPIEPKARQ ELPTQLALEI IVRPAKGVAT TREKVLEDKL RAVLTPLITR HCYPRQLCQ ITCQILESGE DEAEFSLREL SCCINAAFLA LVDAGIALNS MCASIPIAII KDTSDIIVDP TAEQLKISLS V HTLALEFV ...String: MSVQAEIGIL DHVDGSSEFV SQDTKVICSV TGPIEPKARQ ELPTQLALEI IVRPAKGVAT TREKVLEDKL RAVLTPLITR HCYPRQLCQ ITCQILESGE DEAEFSLREL SCCINAAFLA LVDAGIALNS MCASIPIAII KDTSDIIVDP TAEQLKISLS V HTLALEFV NGGKVVKNVL LLDSNGDFNE DQLFSLLELG EQKCQELVTN IRRIIQDNIS PRLVV |
+Macromolecule #73: Exosome complex component RRP42
| Macromolecule | Name: Exosome complex component RRP42 / type: protein_or_peptide / ID: 73 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 29.294398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GHMSLSVAEK SYLYDSLAST PSIRPDGRLP HQFRPIEIFT DFLPSSNGSS RIIASDGSEC IVSIKSKVVD HHVENELLQV DVDIAGQRD DALVVETITS LLNKVLKSGS GVDSSKLQLT KKYSFKIFVD VLVISSHSHP ISLISFAIYS ALNSTYLPKL I SAFDDLEV ...String: GHMSLSVAEK SYLYDSLAST PSIRPDGRLP HQFRPIEIFT DFLPSSNGSS RIIASDGSEC IVSIKSKVVD HHVENELLQV DVDIAGQRD DALVVETITS LLNKVLKSGS GVDSSKLQLT KKYSFKIFVD VLVISSHSHP ISLISFAIYS ALNSTYLPKL I SAFDDLEV EELPTFHDYD MVKLDINPPL VFILAVVGNN MLLDPAANES EVANNGLIIS WSNGKITSPI RSVALNDSNV KS FKPHLLK QGLAMVEKYA PDVVRSLENL |
+Macromolecule #74: Exosome complex component MTR3
| Macromolecule | Name: Exosome complex component MTR3 / type: protein_or_peptide / ID: 74 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.589961 KDa |
| Sequence | String: MNVQDRRRLL GPAAAKPMAF SNTTTHVPEK KSTDLTPKGN ESEQELSLHT GFIENCNGSA LVEARSLGHQ TSLISAVYGP RSIRGSFTS QGTISIQLKN GLLEKYNTNE LKEVSSFLMG IFNSVVNLSR YPKSGIDIFV YLTYDKDLTN NPQDDDSQSK M MSSQISSL ...String: MNVQDRRRLL GPAAAKPMAF SNTTTHVPEK KSTDLTPKGN ESEQELSLHT GFIENCNGSA LVEARSLGHQ TSLISAVYGP RSIRGSFTS QGTISIQLKN GLLEKYNTNE LKEVSSFLMG IFNSVVNLSR YPKSGIDIFV YLTYDKDLTN NPQDDDSQSK M MSSQISSL IPHCITSITL ALADAGIELV DMAGAGEANG TVVSFIKNGE EIVGFWKDDG DDEDLLECLD RCKEQYNRYR DL MISCLMN QET |
+Macromolecule #75: Exosome complex component RRP40
| Macromolecule | Name: Exosome complex component RRP40 / type: protein_or_peptide / ID: 75 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.583354 KDa |
| Sequence | String: MSTFIFPGDS FPVDPTTPVK LGPGIYCDPN TQEIRPVNTG VLHVSAKGKS GVQTAYIDYS SKRYIPSVND FVIGVIIGTF SDSYKVSLQ NFSSSVSLSY MAFPNASKKN RPTLQVGDLV YARVCTAEKE LEAEIECFDS TTGRDAGFGI LEDGMIIDVN L NFARQLLF ...String: MSTFIFPGDS FPVDPTTPVK LGPGIYCDPN TQEIRPVNTG VLHVSAKGKS GVQTAYIDYS SKRYIPSVND FVIGVIIGTF SDSYKVSLQ NFSSSVSLSY MAFPNASKKN RPTLQVGDLV YARVCTAEKE LEAEIECFDS TTGRDAGFGI LEDGMIIDVN L NFARQLLF NNDFPLLKVL AAHTKFEVAI GLNGKIWVKC EELSNTLACY RTIMECCQKN DTAAFKDIAK RQFKEILTVK EE |
+Macromolecule #76: Exosome complex component RRP4
| Macromolecule | Name: Exosome complex component RRP4 / type: protein_or_peptide / ID: 76 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.470176 KDa |
| Sequence | String: MSEVITITKR NGAFQNSSNL SYNNTGISDD ENDEEDIYMH DVNSASKSES DSQIVTPGEL VTDDPIWMRG HGTYFLDNMT YSSVAGTVS RVNRLLSVIP LKGRYAPETG DHVVGRIAEV GNKRWKVDIG GKQHAVLMLG SVNLPGGILR RKSESDELQM R SFLKEGDL ...String: MSEVITITKR NGAFQNSSNL SYNNTGISDD ENDEEDIYMH DVNSASKSES DSQIVTPGEL VTDDPIWMRG HGTYFLDNMT YSSVAGTVS RVNRLLSVIP LKGRYAPETG DHVVGRIAEV GNKRWKVDIG GKQHAVLMLG SVNLPGGILR RKSESDELQM R SFLKEGDL LNAEVQSLFQ DGSASLHTRS LKYGKLRNGM FCQVPSSLIV RAKNHTHNLP GNITVVLGVN GYIWLRKTSQ MD LARDTPS ANNSSSIKST GPTGAVSLNP SITRLEEESS WQIYSDENDP SISNNIRQAI CRYANVIKAL AFCEIGITQQ RIV SAYEAS MVYSNVGELI EKNVMESIGS DILTAEKMRG NGN |
+Macromolecule #77: Exosome complex component CSL4
| Macromolecule | Name: Exosome complex component CSL4 / type: protein_or_peptide / ID: 77 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.619279 KDa |
| Sequence | String: MACNFQFPEI AYPGKLICPQ YGTENKDGED IIFNYVPGPG TKLIQYEHNG RTLEAITATL VGTVRCEEEK KTDQEEEREG TDQSTEEEK SVDASPNDVT RRTVKNILVS VLPGTEKGRK TNKYANNDFA NNLPKEGDIV LTRVTRLSLQ RANVEILAVE D KPSPIDSG ...String: MACNFQFPEI AYPGKLICPQ YGTENKDGED IIFNYVPGPG TKLIQYEHNG RTLEAITATL VGTVRCEEEK KTDQEEEREG TDQSTEEEK SVDASPNDVT RRTVKNILVS VLPGTEKGRK TNKYANNDFA NNLPKEGDIV LTRVTRLSLQ RANVEILAVE D KPSPIDSG IGSNGSGIVA AGGGSGAATF SVSQASSDLG ETFRGIIRSQ DVRSTDRDRV KVIECFKPGD IVRAQVLSLG DG TNYYLTT ARNDLGVVFA RAANGAGGLM YATDWQMMTS PVTGATEKRK CAKPF |
+Macromolecule #78: Exosome complex exonuclease DIS3
| Macromolecule | Name: Exosome complex exonuclease DIS3 / type: protein_or_peptide / ID: 78 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 113.855766 KDa |
| Sequence | String: MSVPAIAPRR KRLADGLSVT QKVFVRSRNG GATKIVREHY LRSDIPCLSR SCTKCPQIVV PDAQNELPKF ILSDSPLELS APIGKHYVV LDTNVVLQAI DLLENPNCFF DVIVPQIVLD EVRNKSYPVY TRLRTLCRDS DDHKRFIVFH NEFSEHTFVE R LPNETIND ...String: MSVPAIAPRR KRLADGLSVT QKVFVRSRNG GATKIVREHY LRSDIPCLSR SCTKCPQIVV PDAQNELPKF ILSDSPLELS APIGKHYVV LDTNVVLQAI DLLENPNCFF DVIVPQIVLD EVRNKSYPVY TRLRTLCRDS DDHKRFIVFH NEFSEHTFVE R LPNETIND RNNRAIRKTC QWYSEHLKPY DINVVLVTND RLNREAATKE VESNIITKSL VQYIELLPNA DDIRDSIPQM DS FDKDLER DTFSDFTFPE YYSTARVMGG LKNGVLYQGN IQISEYNFLE GSVSLPRFSK PVLIVGQKNL NRAFNGDQVI VEL LPQSEW KAPSSIVLDS EHFDVNDNPD IEAGDDDDNN ESSSNTTVIS DKQRRLLAKD AMIAQRSKKI QPTAKVVYIQ RRSW RQYVG QLAPSSVDPQ SSSTQNVFVI LMDKCLPKVR IRTRRAAELL DKRIVISIDS WPTTHKYPLG HFVRDLGTIE SAQAE TEAL LLEHDVEYRP FSKKVLECLP AEGHDWKAPT KLDDPEAVSK DPLLTKRKDL RDKLICSIDP PGCVDINDAL HAKKLP NGN WEVGVHIADV THFVKPGTAL DAEGAARGTS VYLVDKRIDM LPMLLGTDLC SLKPYVDRFA FSVIWELDDS ANIVNVN FM KSVIRSREAF SYEQAQLRID DKTQNDELTM GMRALLKLSV KLKQKRLEAG ALNLASPEVK VHMDSETSDP NEVEIKKL L ATNSLVEEFM LLANISVARK IYDAFPQTAM LRRHAAPPST NFEILNEMLN TRKNMSISLE SSKALADSLD RCVDPEDPY FNTLVRIMST RCMMAAQYFY SGAYSYPDFR HYGLAVDIYT HFTSPIRRYC DVVAHRQLAG AIGYEPLSLT HRDKNKMDMI CRNINRKHR NAQFAGRASI EYYVGQVMRN NESTETGYVI KVFNNGIVVL VPKFGVEGLI RLDNLTEDPN SAAFDEVEYK L TFVPTNSD KPRDVYVFDK VEVQVRSVMD PITSKRKAEL LLK |
+Macromolecule #79: ATP-dependent RNA helicase DOB1
| Macromolecule | Name: ATP-dependent RNA helicase DOB1 / type: protein_or_peptide / ID: 79 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 122.260094 KDa |
| Sequence | String: MDSTDLFDVF EETPVELPTD SNGEKNADTN VGDTPDHTQD KKHGLEEEKE EHEENNSENK KIKSNKSKTE DKNKKVVVPM LADSFEQEA SREVDASKGL TNSETLQVEQ DGKVRLSHQV RHQVALPPNY DYTPIAEHKR VNEARTYPFT LDPFQDTAIS C IDRGESVL ...String: MDSTDLFDVF EETPVELPTD SNGEKNADTN VGDTPDHTQD KKHGLEEEKE EHEENNSENK KIKSNKSKTE DKNKKVVVPM LADSFEQEA SREVDASKGL TNSETLQVEQ DGKVRLSHQV RHQVALPPNY DYTPIAEHKR VNEARTYPFT LDPFQDTAIS C IDRGESVL VSAHTSAGKT VVAEYAIAQS LKNKQRVIYT SPIKALSNQK YRELLAEFGD VGLMTGDITI NPDAGCLVMT TE ILRSMLY RGSEVMREVA WVIFDEVHYM RDKERGVVWE ETIILLPDKV RYVFLSATIP NAMEFAEWIC KIHSQPCHIV YTN FRPTPL QHYLFPAHGD GIYLVVDEKS TFREENFQKA MASISNQIGD DPNSTDSRGK KGQTYKGGSA KGDAKGDIYK IVKM IWKKK YNPVIVFSFS KRDCEELALK MSKLDFNSDD EKEALTKIFN NAIALLPETD RELPQIKHIL PLLRRGIGIH HSGLL PILK EVIEILFQEG FLKVLFATET FSIGLNMPAK TVVFTSVRKW DGQQFRWVSG GEYIQMSGRA GRRGLDDRGI VIMMID EKM EPQVAKGMVK GQADRLDSAF HLGYNMILNL MRVEGISPEF MLEHSFFQFQ NVISVPVMEK KLAELKKDFD GIEVEDE EN VKEYHEIEQA IKGYREDVRQ VVTHPANALS FLQPGRLVEI SVNGKDNYGW GAVVDFAKRI NKRNPSAVYT DHESYIVN V VVNTMYIDSP VNLLKPFNPT LPEGIRPAEE GEKSICAVIP ITLDSIKSIG NLRLYMPKDI RASGQKETVG KSLREVNRR FPDGIPVLDP VKNMKIEDED FLKLMKKIDV LNTKLSSNPL TNSMRLEELY GKYSRKHDLH EDMKQLKRKI SESQAVIQLD DLRRRKRVL RRLGFCTPND IIELKGRVAC EISSGDELLL TELIFNGNFN ELKPEQAAAL LSCFAFQERC KEAPRLKPEL A EPLKAMRE IAAKIAKIMK DSKIEVVEKD YVESFRHELM EVVYEWCRGA TFTQICKMTD VYEGSLIRMF KRLEELVKEL VD VANTIGN SSLKEKMEAV LKLIHRDIVS AGSLYL |
+Macromolecule #65: 5'ETS RNA
| Macromolecule | Name: 5'ETS RNA / type: rna / ID: 65 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 225.543094 KDa |
| Sequence | String: AUGCGAAAGC AGUUGAAGAC AAGUUCGAAA AGAGUUUGGA AACGAAUUCG AGUAGGCUUG UCGUUCGUUA UGUUUUUGUA AAUGGCCUC GUCAAACGGU GGAGAGAGUC GCUAGGUGAU CGUCAGAUCU GCCUAGUCUC UAUACAGCGU GUUUAAUUGA C AUGGGUUG ...String: AUGCGAAAGC AGUUGAAGAC AAGUUCGAAA AGAGUUUGGA AACGAAUUCG AGUAGGCUUG UCGUUCGUUA UGUUUUUGUA AAUGGCCUC GUCAAACGGU GGAGAGAGUC GCUAGGUGAU CGUCAGAUCU GCCUAGUCUC UAUACAGCGU GUUUAAUUGA C AUGGGUUG AUGCGUAUUG AGAGAUACAA UUUGGGAAGA AAUUCCCAGA GUGUGUUUCU UUUGCGUUUA ACCUGAACAG UC UCAUCGU GGGCAUCUUG CGAUUCCAUU GGUGAGCAGC GAAGGAUUUG GUGGAUUACU AGCUAAUAGC AAUCUAUUUC AAA GAAUUC AAACUUGGGG GAAUGCCUUG UUGAAUAGCC GGUCGCAAGA CUGUGAUUCU UCAAGUGUAA CCUCCUCUCA AAUC AGCGA UAUCAAACGU ACCAUUCCGU GAAACACCGG GGUAUCUGUU UGGUGGAACC UGAUUAGAGG AAACUCAAAG AGUGC UAUG GUAUGGUGAC GGAGUGCGCU GGUCAAGAGU GUAAAAGCUU UUUGAACAGA GAGCAUUUCC GGCAGCAGAG AGACCU GAA AAAGCAAUUU UUCUGGAAUU UCAGCUGUUU CCAAACUCAA UAAGUAUCUU CUAGCAAGAG GGAAUAGGUG GGAAAAA AA AAAAGAGAUU UCGGUUUCUU UCUUUUUUAC UGCUUGUUGC UUCUUCUUUU AAGAUAGU |
+Macromolecule #66: 18S rRNA
| Macromolecule | Name: 18S rRNA / type: rna / ID: 66 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 582.381688 KDa |
| Sequence | String: AAGAUAGUUA UCUGGUUGAU CCUGCCAGUA GUCAUAUGCU UGUCUCAAAG AUUAAGCCAU GCAUGUCUAA GUAUAAGCAA UUUAUACAG UGAAACUGCG AAUGGCUCAU UAAAUCAGUU AUCGUUUAUU UGAUAGUUCC UUUACUACAU GGUAUAACUG U GGUAAUUC ...String: AAGAUAGUUA UCUGGUUGAU CCUGCCAGUA GUCAUAUGCU UGUCUCAAAG AUUAAGCCAU GCAUGUCUAA GUAUAAGCAA UUUAUACAG UGAAACUGCG AAUGGCUCAU UAAAUCAGUU AUCGUUUAUU UGAUAGUUCC UUUACUACAU GGUAUAACUG U GGUAAUUC UAGAGCUAAU ACAUGCUUAA AAUCUCGACC CUUUGGAAGA GAUGUAUUUA UUAGAUAAAA AAUCAAUGUC UU CGGACUC UUUGAUGAUU CAUAAUAACU UUUCGAAUCG CAUGGCCUUG UGCUGGCGAU GGUUCAUUCA AAUUUCUGCC CUA UCAACU UUCGAUGGUA GGAUAGUGGC CUACCAUGGU UUCAACGGGU AACGGGGAAU AAGGGUUCGA UUCCGGAGAG GGAG CCUGA GAAACGGCUA CCACAUCCAA GGAAGGCAGC AGGCGCGCAA AUUACCCAAU CCUAAUUCAG GGAGGUAGUG ACAAU AAAU AACGAUACAG GGCCCAUUCG GGUCUUGUAA UUGGAAUGAG UACAAUGUAA AUACCUUAAC GAGGAACAAU UGGAGG GCA AGUCUGGUGC CAGCAGCCGC GGUAAUUCCA GCUCCAAUAG CGUAUAUUAA AGUUGUUGCA GUUAAAAAGC UCGUAGU UG AACUUUGGGC CCGGUUGGCC GGUCCGAUUU UUUCGUGUAC UGGAUUUCCA ACGGGGCCUU UCCUUCUGGC UAACCUUG A GUCCUUGUGG CUCUUGGCGA ACCAGGACUU UUACUUUGAA AAAAUUAGAG UGUUCAAAGC AGGCGUAUUG CUCGAAUAU AUUAGCAUGG AAUAAUAGAA UAGGACGUUU GGUUCUAUUU UGUUGGUUUC UAGGACCAUC GUAAUGAUUA AUAGGGACGG UCGGGGGCA UCAGUAUUCA AUUGUCAGAG GUGAAAUUCU UGGAUUUAUU GAAGACUAAC UACUGCGAAA GCAUUUGCCA A GGACGUUU UCAUUAAUCA AGAACGAAAG UUAGGGGAUC GAAGAUGAUC AGAUACCGUC GUAGUCUUAA CCAUAAACUA UG CCGACUA GGGAUCGGGU GGUGUUUUUU UAAUGACCCA CUCGGCACCU UACGAGAAAU CAAAGUCUUU GGGUUCUGGG GGG AGUAUG GUCGCAAGGC UGAAACUUAA AGGAAUUGAC GGAAGGGCAC CACCAGGAGU GGAGCCUGCG GCUUAAUUUG ACUC AACAC GGGGAAACUC ACCAGGUCCA GACACAAUAA GGAUUGACAG AUUGAGAGCU CUUUCUUGAU UUUGUGGGUG GUGGU GCAU GGCCGUUCUU AGUUGGUGGA GUGAUUUGUC UGCUUAAUUG CGAUAACGAA CGAGACCUUA ACCUACUAAA UAGUGG UGC UAGCAUUUGC UGGUUAUCCA CUUCUUAGAG GGACUAUCGG UUUCAAGCCG AUGGAAGUUU GAGGCAAUAA CAGGUCU GU GAUGCCCUUA GACGUUCUGG GCCGCACGCG CGCUACACUG ACGGAGCCAG CGAGUCUAAC CUUGGCCGAG AGGUCUUG G UAAUCUUGUG AAACUCCGUC GUGCUGGGGA UAGAGCAUUG UAAUUAUUGC UCUUCAACGA GGAAUUCCUA GUAAGCGCA AGUCAUCAGC UUGCGUUGAU UACGUCCCUG CCCUUUGUAC ACACCGCCCG UCGCUAGUAC CGAUUGAAUG GCUUAGUGAG GCCUCAGGA UCUGCUUAGA GAAGGGGGCA ACUCCAUCUC AGAGCGGAGA AUUUGGACAA ACUUGGUCAU UUAGAGGAAC U AAAAGUCG UAACAAGGUU UCCGUAGGUG AACCUGCGGA AGGAUCAUUA |
+Macromolecule #67: U3 snoRNA
| Macromolecule | Name: U3 snoRNA / type: rna / ID: 67 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 106.503258 KDa |
| Sequence | String: GUCGACGUAC UUCAUAGGAU CAUUUCUAUA GGAAUCGUCA CUCUUUGACU CUUCAAAAGA GCCACUGAAU CCAACUUGGU UGAUGAGUC CCAUAACCUU UGUACCCCAG AGUGAGAAAC CGAAAUUGAA UCUAAAUUAG CUUGGUCCGC AAUCCUUAGC G GUUCGGCC ...String: GUCGACGUAC UUCAUAGGAU CAUUUCUAUA GGAAUCGUCA CUCUUUGACU CUUCAAAAGA GCCACUGAAU CCAACUUGGU UGAUGAGUC CCAUAACCUU UGUACCCCAG AGUGAGAAAC CGAAAUUGAA UCUAAAUUAG CUUGGUCCGC AAUCCUUAGC G GUUCGGCC AUCUAUAAUU UUGAAUAAAA AUUUUGCUUU GCCGUUGCAU UUGUAGUUUU UUCCUUUGGA AGUAAUUACA AU AUUUUAU GGCGCGAUGA UCUUGACCCA UCCUAUGUAC UUCUUUUUUG AAGGGAUAGG GCUCUAUGGG UGGGUACAAA UGG CAGUCU GACAAGU |
+Macromolecule #68: RNA
| Macromolecule | Name: RNA / type: rna / ID: 68 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 6.851971 KDa |
| Sequence | String: AAAAUUUAAA UUUUUUUUUU UU |
+Macromolecule #80: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 80 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
+Macromolecule #81: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 81 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
+Macromolecule #82: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 82 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller




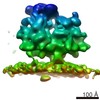

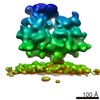
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)