[English] 日本語
 Yorodumi
Yorodumi- EMDB-10789: Structure of ExbB pentamer from Serratia marcescens by single par... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10789 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of ExbB pentamer from Serratia marcescens by single particle cryo electron microscopy | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / iron uptake / proton transfer / TonB complex / METAL TRANSPORT | |||||||||
| Function / homology | TonB-system energizer ExbB type-1 / : / MotA/TolQ/ExbB proton channel / MotA/TolQ/ExbB proton channel family / protein import / transmembrane transporter activity / plasma membrane / Biopolymer transport protein ExbB Function and homology information Function and homology information | |||||||||
| Biological species |  Serratia marcescens (bacteria) Serratia marcescens (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Biou V / Delepelaire P | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation | Journal: J Struct Biol / Year: 2012 Title: RELION: implementation of a Bayesian approach to cryo-EM structure determination. Authors: Sjors H W Scheres /  Abstract: RELION, for REgularized LIkelihood OptimizatioN, is an open-source computer program for the refinement of macromolecular structures by single-particle analysis of electron cryo-microscopy (cryo-EM) ...RELION, for REgularized LIkelihood OptimizatioN, is an open-source computer program for the refinement of macromolecular structures by single-particle analysis of electron cryo-microscopy (cryo-EM) data. Whereas alternative approaches often rely on user expertise for the tuning of parameters, RELION uses a Bayesian approach to infer parameters of a statistical model from the data. This paper describes developments that reduce the computational costs of the underlying maximum a posteriori (MAP) algorithm, as well as statistical considerations that yield new insights into the accuracy with which the relative orientations of individual particles may be determined. A so-called gold-standard Fourier shell correlation (FSC) procedure to prevent overfitting is also described. The resulting implementation yields high-quality reconstructions and reliable resolution estimates with minimal user intervention and at acceptable computational costs. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Functional and structural characterization of Serratia marcescens ExbB: determinants of the interaction with HasB/TonB Authors: Biou V / Chami M / Coureux PD / Laurent B / Ntsogo Y / Izadi-Pruneyre N / Malosse C / Chamot-Rooke J / Stahlberg H / Delepelaire P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10789.map.gz emd_10789.map.gz | 13.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10789-v30.xml emd-10789-v30.xml emd-10789.xml emd-10789.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10789_fsc.xml emd_10789_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_10789.png emd_10789.png | 132.3 KB | ||
| Masks |  emd_10789_msk_1.map emd_10789_msk_1.map | 184 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10789.cif.gz emd-10789.cif.gz | 7.2 KB | ||
| Others |  emd_10789_additional_1.map.gz emd_10789_additional_1.map.gz emd_10789_half_map_1.map.gz emd_10789_half_map_1.map.gz emd_10789_half_map_2.map.gz emd_10789_half_map_2.map.gz | 3.6 MB 145.7 MB 145.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10789 http://ftp.pdbj.org/pub/emdb/structures/EMD-10789 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10789 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10789 | HTTPS FTP |
-Related structure data
| Related structure data |  6ye4MC  7ajqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10789.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10789.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
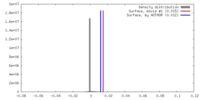
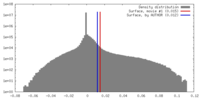
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10789_msk_1.map emd_10789_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_10789_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10789_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10789_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homopentamer of ExbB
| Entire | Name: Homopentamer of ExbB |
|---|---|
| Components |
|
-Supramolecule #1: Homopentamer of ExbB
| Supramolecule | Name: Homopentamer of ExbB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: the expressed sequence corresponds to the mature sequence after signal peptide cleavage. |
|---|---|
| Source (natural) | Organism:  Serratia marcescens (bacteria) / Strain: Db11 Serratia marcescens (bacteria) / Strain: Db11 |
| Molecular weight | Theoretical: 174 KDa |
-Macromolecule #1: Biopolymer transport protein ExbB
| Macromolecule | Name: Biopolymer transport protein ExbB / type: protein_or_peptide / ID: 1 / Details: a 6-histidine tag is present at the C-terminus / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Serratia marcescens (bacteria) Serratia marcescens (bacteria) |
| Molecular weight | Theoretical: 30.413621 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: APAANPAVTE SVAPTTAPAP AAAAPESITP VNPAPTIQPP ETRGMDLSIW GMYQHADAVV KAVMIGLVLA SIVTWTILFA KGSELLRAK RRLRREQLAL AEARSLDEAS ELAQNFSPES VSAVLLNDAQ NELELSAESN DNNGIKERTG FRLERRVAAY S RNMGRGNG ...String: APAANPAVTE SVAPTTAPAP AAAAPESITP VNPAPTIQPP ETRGMDLSIW GMYQHADAVV KAVMIGLVLA SIVTWTILFA KGSELLRAK RRLRREQLAL AEARSLDEAS ELAQNFSPES VSAVLLNDAQ NELELSAESN DNNGIKERTG FRLERRVAAY S RNMGRGNG FLATIGAISP FVGLFGTVWG IMNSFIGIAH SQTTNLAVVA PGIAEALLAT AMGLVAAIPA VVIYNIFARV IS GHRAQVG DVAAQVLLLQ GRDLDLAATA EAKRSQHAHQ LRAGHHHHHH UniProtKB: Biopolymer transport protein ExbB |
-Macromolecule #2: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 2 / Number of copies: 5 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20mM Tris-HCl pH 8,0 100mM NaCl 0,0015% LMNG | ||||||||||||
| Grid | Model: Quantifoil R2/4 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA EM CPC | ||||||||||||
| Details | the sample was monodisperse as evidenced by gel filtration column |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 83.0 K / Max: 93.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-56 / Number grids imaged: 1 / Number real images: 3122 / Average exposure time: 7.0 sec. / Average electron dose: 55.95 e/Å2 Details: frames were weighted according to electron dose and particle movement during Relion bayesian polishing procedure. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 10-234 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | the ExbB sequence from S. marcescens was modeled by homology from the 5SV0 monomer from E. coli using Phyre software and the pentamer was generated using the 5SV0 symmetry. real space refinement was carried out with rigid body, simulated annealing and morphing steps. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 55.3 / Target criteria: Correlation coefficient |
| Output model |  PDB-6ye4: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)