+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11806 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of ExbBD from Serratia Marcescens | |||||||||
 Map data Map data | Cryosparc sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TonB transport / MEMBRANE PROTEIN / IRON UPTAKE / PROTON TRANSFER / TONB COMPLEX / METAL TRANSPORT / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransmembrane transporter activity / membrane => GO:0016020 / protein transport / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Serratia marcescens (bacteria) Serratia marcescens (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Biou V / Adaixo R | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog. Authors: Valérie Biou / Ricardo Jorge Diogo Adaixo / Mohamed Chami / Pierre-Damien Coureux / Benoist Laurent / Véronique Yvette Ntsogo Enguéné / Gisele Cardoso de Amorim / Nadia Izadi-Pruneyre / ...Authors: Valérie Biou / Ricardo Jorge Diogo Adaixo / Mohamed Chami / Pierre-Damien Coureux / Benoist Laurent / Véronique Yvette Ntsogo Enguéné / Gisele Cardoso de Amorim / Nadia Izadi-Pruneyre / Christian Malosse / Julia Chamot-Rooke / Henning Stahlberg / Philippe Delepelaire /    Abstract: ExbB and ExbD are cytoplasmic membrane proteins that associate with TonB to convey the energy of the proton-motive force to outer membrane receptors in Gram-negative bacteria for iron uptake. The ...ExbB and ExbD are cytoplasmic membrane proteins that associate with TonB to convey the energy of the proton-motive force to outer membrane receptors in Gram-negative bacteria for iron uptake. The opportunistic pathogen Serratia marcescens (Sm) possesses both TonB and a heme-specific TonB paralog, HasB. ExbB has a long periplasmic extension absent in other bacteria such as E. coli (Ec). Long ExbB's are found in several genera of Alphaproteobacteria, most often in correlation with a hasB gene. We investigated specificity determinants of ExbB and HasB. We determined the cryo-EM structures of ExbB and of the ExbB-ExbD complex from S. marcescens. ExbB alone is a stable pentamer, and its complex includes two ExbD monomers. We showed that ExbB extension interacts with HasB and is involved in heme acquisition and we identified key residues in the membrane domain of ExbB and ExbB, essential for function and likely involved in the interaction with TonB/HasB. Our results shed light on the class of inner membrane energy machinery formed by ExbB, ExbD and HasB. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Functional and structural characterization of Serratia marcescens ExbB: determinants of the interaction with HasB/TonB Authors: Biou V / Chami M / Coureux PD / Laurent B / Ntsogo Y / Izadi-Pruneyre N / Malosse C / Chamot-Rooke J / Stahlberg H / Delepelaire P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11806.map.gz emd_11806.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11806-v30.xml emd-11806-v30.xml emd-11806.xml emd-11806.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11806_fsc.xml emd_11806_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11806.png emd_11806.png | 66.2 KB | ||
| Filedesc metadata |  emd-11806.cif.gz emd-11806.cif.gz | 6.6 KB | ||
| Others |  emd_11806_half_map_1.map.gz emd_11806_half_map_1.map.gz emd_11806_half_map_2.map.gz emd_11806_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11806 http://ftp.pdbj.org/pub/emdb/structures/EMD-11806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11806 | HTTPS FTP |
-Related structure data
| Related structure data |  7ajqMC  6ye4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11806.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11806.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
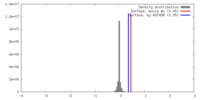
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map B
| File | emd_11806_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
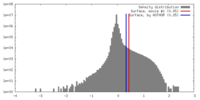
| Density Histograms |
-Half map: half map A
| File | emd_11806_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 5-ExbB + 2-ExbD complex
| Entire | Name: 5-ExbB + 2-ExbD complex |
|---|---|
| Components |
|
-Supramolecule #1: 5-ExbB + 2-ExbD complex
| Supramolecule | Name: 5-ExbB + 2-ExbD complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Serratia marcescens (bacteria) Serratia marcescens (bacteria) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Biopolymer transport protein ExbB
| Macromolecule | Name: Biopolymer transport protein ExbB / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Serratia marcescens (bacteria) Serratia marcescens (bacteria) |
| Molecular weight | Theoretical: 29.584752 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: APAANPAVTE SVAPTTAPAP AAAAPESITP VNPAPTIQPP ETRGMDLSIW GMYQHADAVV KAVMIGLVLA SIVTWTILFA KGSELLRAK RRLRREQLAL AEARSLDEAS ELAQNFSPES VSAVLLNDAQ NELELSAESN DNNGIKERTG FRLERRVAAY S RNMGRGNG ...String: APAANPAVTE SVAPTTAPAP AAAAPESITP VNPAPTIQPP ETRGMDLSIW GMYQHADAVV KAVMIGLVLA SIVTWTILFA KGSELLRAK RRLRREQLAL AEARSLDEAS ELAQNFSPES VSAVLLNDAQ NELELSAESN DNNGIKERTG FRLERRVAAY S RNMGRGNG FLATIGAISP FVGLFGTVWG IMNSFIGIAH SQTTNLAVVA PGIAEALLAT AMGLVAAIPA VVIYNIFARV IS GHRAQVG DVAAQVLLLQ GRDLDLAATA EAKRSQHAHQ LRAG UniProtKB: Biopolymer transport protein ExbB |
-Macromolecule #2: Biopolymer transport protein ExbD
| Macromolecule | Name: Biopolymer transport protein ExbD / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Serratia marcescens (bacteria) Serratia marcescens (bacteria) |
| Molecular weight | Theoretical: 16.26869 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAMRLNEDLD DSGELHEINV TPFIDVMLVL LIIFMVAAPL ATVDIRVDLP ASSAKPQPRP EKPVFLSVKA DKQLYVGDQP VNADQLTSV LDQRTQANKE TTIFFQADKS VDYETLMSVM DTLRKAGYLK VGLVGMEGAA KHHHHHH UniProtKB: Biopolymer transport protein ExbD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: C-flat / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK II | ||||||||||||
| Details | the sample was monodisperse as seen on gel filtration chromatogram |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-48 / Number grids imaged: 1 / Number real images: 4000 / Average exposure time: 12.0 sec. / Average electron dose: 4.58 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.8000000000000003 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 139000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 169 / Target criteria: correlation coefficient |
| Output model |  PDB-7ajq: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)