[English] 日本語
 Yorodumi
Yorodumi- EMDB-10528: Structure of the chaperonin gp146 from the bacteriophage EL (Pseu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10528 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in the apo state | |||||||||
 Map data Map data | CryoEM map of apo gp146, the chaperonin from bacteriophage EL (Pseudomonas putida) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | molecular chaperone / ATPase / chaperonin / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-dependent protein folding chaperone / protein refolding / ATP binding / metal ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Pseudomonas phage EL (virus) Pseudomonas phage EL (virus) | |||||||||
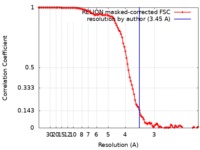
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Bracher A / Wang H | |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2020 Journal: PLoS One / Year: 2020Title: Structure and conformational cycle of a bacteriophage-encoded chaperonin. Authors: Andreas Bracher / Simanta S Paul / Huping Wang / Nadine Wischnewski / F Ulrich Hartl / Manajit Hayer-Hartl /  Abstract: Chaperonins are ubiquitous molecular chaperones found in all domains of life. They form ring-shaped complexes that assist in the folding of substrate proteins in an ATP-dependent reaction cycle. Key ...Chaperonins are ubiquitous molecular chaperones found in all domains of life. They form ring-shaped complexes that assist in the folding of substrate proteins in an ATP-dependent reaction cycle. Key to the folding cycle is the transient encapsulation of substrate proteins by the chaperonin. Here we present a structural and functional characterization of the chaperonin gp146 (ɸEL) from the phage EL of Pseudomonas aeruginosa. ɸEL, an evolutionarily distant homolog of bacterial GroEL, is active in ATP hydrolysis and prevents the aggregation of denatured protein in a nucleotide-dependent manner. However, ɸEL failed to refold the encapsulation-dependent model substrate rhodanese and did not interact with E. coli GroES, the lid-shaped co-chaperone of GroEL. ɸEL forms tetradecameric double-ring complexes, which dissociate into single rings in the presence of ATP. Crystal structures of ɸEL (at 3.54 and 4.03 Å) in presence of ATP•BeFx revealed two distinct single-ring conformational states, both with open access to the ring cavity. One state showed uniform ATP-bound subunit conformations (symmetric state), whereas the second combined distinct ATP- and ADP-bound subunit conformations (asymmetric state). Cryo-electron microscopy of apo-ɸEL revealed a double-ring structure composed of rings in the asymmetric state (3.45 Å resolution). We propose that the phage chaperonin undergoes nucleotide-dependent conformational switching between double- and single rings and functions in aggregation prevention without substrate protein encapsulation. Thus, ɸEL may represent an evolutionarily more ancient chaperonin prior to acquisition of the encapsulation mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10528.map.gz emd_10528.map.gz | 12.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10528-v30.xml emd-10528-v30.xml emd-10528.xml emd-10528.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10528_fsc.xml emd_10528_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_10528.png emd_10528.png | 91.6 KB | ||
| Filedesc metadata |  emd-10528.cif.gz emd-10528.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10528 http://ftp.pdbj.org/pub/emdb/structures/EMD-10528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10528 | HTTPS FTP |
-Validation report
| Summary document |  emd_10528_validation.pdf.gz emd_10528_validation.pdf.gz | 446.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10528_full_validation.pdf.gz emd_10528_full_validation.pdf.gz | 446 KB | Display | |
| Data in XML |  emd_10528_validation.xml.gz emd_10528_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_10528_validation.cif.gz emd_10528_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10528 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10528 | HTTPS FTP |
-Related structure data
| Related structure data |  6tmvMC  6tmtC  6tmuC  6tmwC  6tmxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10528.map.gz / Format: CCP4 / Size: 96.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10528.map.gz / Format: CCP4 / Size: 96.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of apo gp146, the chaperonin from bacteriophage EL (Pseudomonas putida) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The chaperonin gp146 from the bacteriophage EL (Pseudomonas aerug...
| Entire | Name: The chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in the apo state |
|---|---|
| Components |
|
-Supramolecule #1: The chaperonin gp146 from the bacteriophage EL (Pseudomonas aerug...
| Supramolecule | Name: The chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in the apo state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage EL (virus) Pseudomonas phage EL (virus) |
| Molecular weight | Theoretical: 864 KDa |
-Macromolecule #1: Putative GroEL-like chaperonine protein
| Macromolecule | Name: Putative GroEL-like chaperonine protein / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage EL (virus) Pseudomonas phage EL (virus) |
| Molecular weight | Theoretical: 61.760941 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSQTLLVHGK DAQGIIKQVL SEVYDAVTST MGPNGQLVMI KNGVSTKTTK DGVTVARSIR FADEAHELVN RVITEPATKT DEECGDGTT TTIMLTHALY HLFKDFPGFQ HHRNIEDLVE RVIQRLESMA IRVEVDDPRL YQVALTSSNQ DEKLARLVSE L YANNKGSY ...String: MSQTLLVHGK DAQGIIKQVL SEVYDAVTST MGPNGQLVMI KNGVSTKTTK DGVTVARSIR FADEAHELVN RVITEPATKT DEECGDGTT TTIMLTHALY HLFKDFPGFQ HHRNIEDLVE RVIQRLESMA IRVEVDDPRL YQVALTSSNQ DEKLARLVSE L YANNKGSY PDIELKEGVN FEDQIEQTTG RTIRMFYANP WFAKGHQGGV TELTGFTAFV IDRRIDKEDT QKLIDGVNHL VK THKQHLA LPILLIARSF EEAANSTLMQ LNAAHPTLVE DGRPWLIPLS TPVGGAIGTS ELQDIAVMLN APMLSDVADL TKL DTHSIN GQHGQLELGG NRSILKSTTP KDEDRIEQHA RGIEELLEGF SLSDKFSVRA RYNERRIRTL RGKLITISVG GETY SEVKE RVDRYEDVVK AIRSALENGI LPGGGVSLVK AVFGTIKEGL EDKDQSAEFA KRYINSGIAN ELMRLSTIQH KLLFK DTAL YKENGSFHFN DDWLNTPTVM NLATGEIGTP EGLGIYDTAY ASITALKGGL QTAKILATTK TLILGEKLSA VKVR UniProtKB: Putative GroEL-like chaperonine protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.125 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: Blot time was 2 sec.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Number grids imaged: 1 / #0 - Number real images: 2519 / #0 - Average exposure time: 0.11984 sec. / #0 - Average electron dose: 1.552 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Number grids imaged: 1 / #1 - Number real images: 3063 / #1 - Average exposure time: 0.12 sec. / #1 - Average electron dose: 1.55 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Jelly body refinement C2 symmetry NCS restraints |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-6tmv: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)