[English] 日本語
 Yorodumi
Yorodumi- EMDB-5447: Molecular Architecture of the Unliganded Membrane-Bound HIV-1 Env... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5447 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular Architecture of the Unliganded Membrane-Bound HIV-1 Envelope Glycoprotein Trimer | |||||||||
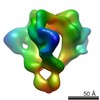
 Map data Map data | Reconstruction of the unliganded HIV-1 envelope glycoprotein trimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / envelope spike / trimer / membrane glycoprotein | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Mao Y / Wang L / Sodroski J | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2013 Journal: Proc Natl Acad Sci U S A / Year: 2013Title: Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Authors: Youdong Mao / Liping Wang / Christopher Gu / Alon Herschhorn / Anik Désormeaux / Andrés Finzi / Shi-Hua Xiang / Joseph G Sodroski /  Abstract: The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) trimer, a membrane-fusing machine, mediates virus entry into host cells and is the sole virus-specific target for ...The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) trimer, a membrane-fusing machine, mediates virus entry into host cells and is the sole virus-specific target for neutralizing antibodies. Binding the receptors, CD4 and CCR5/CXCR4, triggers Env conformational changes from the metastable unliganded state to the fusion-active state. We used cryo-electron microscopy to obtain a 6-Å structure of the membrane-bound, heavily glycosylated HIV-1 Env trimer in its uncleaved and unliganded state. The spatial organization of secondary structure elements reveals that the unliganded conformations of both glycoprotein (gp)120 and gp41 subunits differ from those induced by receptor binding. The gp120 trimer association domains, which contribute to interprotomer contacts in the unliganded Env trimer, undergo rearrangement upon CD4 binding. In the unliganded Env, intersubunit interactions maintain the gp41 ectodomain helical bundles in a "spring-loaded" conformation distinct from the extended helical coils of the fusion-active state. Quaternary structure regulates the virus-neutralizing potency of antibodies targeting the conserved CD4-binding site on gp120. The Env trimer architecture provides mechanistic insights into the metastability of the unliganded state, receptor-induced conformational changes, and quaternary structure-based strategies for immune evasion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5447.map.gz emd_5447.map.gz | 59.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5447-v30.xml emd-5447-v30.xml emd-5447.xml emd-5447.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5447_1.tif emd_5447_1.tif | 248.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5447 http://ftp.pdbj.org/pub/emdb/structures/EMD-5447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5447 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10003 (Title: HIV-1 envelope glycoprotein trimer micrographs / Data size: 586.8 EMPIAR-10003 (Title: HIV-1 envelope glycoprotein trimer micrographs / Data size: 586.8 Data #1: 10 focal pair micrographs of HIV-1 Envelope Glycoprotein Trimer [picked particles - single frame - unprocessed] Data #2: Full set of 4683 focal pair micrographs [micrographs - focal pairs - contrast inverted])  EMPIAR-10007 (Title: Low-contrast particle stack for HIV-1 Env gp160 precursor (Spider stack) EMPIAR-10007 (Title: Low-contrast particle stack for HIV-1 Env gp160 precursor (Spider stack)Data size: 164.2 Data #1: U115 raw particle images [picked particles - multiframe - unprocessed])  EMPIAR-10008 (Title: Low-contrast particle stack for HIV-1 Env gp160 precursor (MRC stack) EMPIAR-10008 (Title: Low-contrast particle stack for HIV-1 Env gp160 precursor (MRC stack)Data size: 240.2 / Data #1: U115 [picked particles - multiframe - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5447.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5447.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the unliganded HIV-1 envelope glycoprotein trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.74688 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
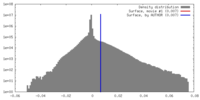
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV-1 envelope glycoprotein trimer
| Entire | Name: HIV-1 envelope glycoprotein trimer |
|---|---|
| Components |
|
-Supramolecule #1000: HIV-1 envelope glycoprotein trimer
| Supramolecule | Name: HIV-1 envelope glycoprotein trimer / type: sample / ID: 1000 Details: The sample was monodisperse and solubilized in Cymal-6. Oligomeric state: three gp120 exterior subunits and three gp41 transmembrane subunits Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 440 KDa / Theoretical: 435 KDa / Method: Sedimentation, gel electrophoresis |
-Macromolecule #1: HIV-1 envelope glycoprotein trimer precursor
| Macromolecule | Name: HIV-1 envelope glycoprotein trimer precursor / type: protein_or_peptide / ID: 1 / Name.synonym: Env trimer, gp160 trimer Details: The glycoprotein was expressed in 293F cells and was purified from the extracted plasma membranes. The cDNA sequence is derived from HIV-1 JR-FL isolate. Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / synonym: HIV-1 Human immunodeficiency virus 1 / synonym: HIV-1 |
| Molecular weight | Experimental: 440 KDa / Theoretical: 435 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant plasmid: pcDNA3.1 in 293F cell line Homo sapiens (human) / Recombinant plasmid: pcDNA3.1 in 293F cell line |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Tris-HCl, 300 mM NaCl and 0.01% Cymal-6 |
| Grid | Details: 200 mesh and 400 mesh C-flat grid with thin holey carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 93 K / Instrument: FEI VITROBOT MARK IV Method: Held for 2 s, blotted by filter papers for 2 s before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 89 K / Max: 93 K / Average: 91 K |
| Date | Sep 1, 2009 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Number real images: 5991 / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 200835 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder: CT3500 / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Wiener filter |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 6.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER Details: The final map was deconvoluted and amplitude-corrected with a B-factor of 250 and was low-pass filtered at 5.6 Angstrom with a cosine edge of 8-Fourier-pixel width. The final resolution of ...Details: The final map was deconvoluted and amplitude-corrected with a B-factor of 250 and was low-pass filtered at 5.6 Angstrom with a cosine edge of 8-Fourier-pixel width. The final resolution of the refined cryo-EM map, measured by FSC-0.5 cutoff, is 6 Angstrom without masking of the background noise in the map and is 5.66 Angstrom with of masking the background noise. Number images used: 582914 |
| Final angle assignment | Details: 0.5 degree of angular step |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: G |
|---|---|
| Software | Name: O, Coot, CNS, Chimera, Modeller |
| Details | Protocol: Flexible fitting |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)