[English] 日本語
 Yorodumi
Yorodumi- EMDB-5418: Subunit organization of the membrane-bound HIV-1 envelope glycopr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5418 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer | |||||||||
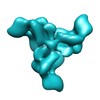
 Map data Map data | Reconstruction of the membrane-bound HIV-1 envelope glycoprotein trimer precursor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / envelope glycoprotein / virus entry | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.8 Å | |||||||||
 Authors Authors | Mao Y / Wang L / Gu C / Herschhorn A / Xiang SH / Haim H / Yang X / Sodroski J | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2012 Journal: Nat Struct Mol Biol / Year: 2012Title: Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Authors: Youdong Mao / Liping Wang / Christopher Gu / Alon Herschhorn / Shi-Hua Xiang / Hillel Haim / Xinzhen Yang / Joseph Sodroski /  Abstract: The trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) spike is a molecular machine that mediates virus entry into host cells and is the sole target for virus- ...The trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) spike is a molecular machine that mediates virus entry into host cells and is the sole target for virus-neutralizing antibodies. The mature Env spike results from cleavage of a trimeric glycoprotein precursor, gp160, into three gp120 and three gp41 subunits. Here, we describe an ~11-Å cryo-EM structure of the trimeric HIV-1 Env precursor in its unliganded state. The three gp120 and three gp41 subunits form a cage-like structure with an interior void surrounding the trimer axis. Interprotomer contacts are limited to the gp41 transmembrane region, the torus-like gp41 ectodomain and a trimer-association domain of gp120 composed of the V1, V2 and V3 variable regions. The cage-like architecture, which is unique among characterized viral envelope proteins, restricts antibody access, reflecting requirements imposed by HIV-1 persistence in the host. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5418.map.gz emd_5418.map.gz | 166.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5418-v30.xml emd-5418-v30.xml emd-5418.xml emd-5418.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5418.jpg emd_5418.jpg | 28.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5418 http://ftp.pdbj.org/pub/emdb/structures/EMD-5418 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5418 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5418 | HTTPS FTP |
-Validation report
| Summary document |  emd_5418_validation.pdf.gz emd_5418_validation.pdf.gz | 79 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5418_full_validation.pdf.gz emd_5418_full_validation.pdf.gz | 78.1 KB | Display | |
| Data in XML |  emd_5418_validation.xml.gz emd_5418_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5418 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5418 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5418 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5418 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5418.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5418.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the membrane-bound HIV-1 envelope glycoprotein trimer precursor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.99 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
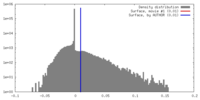
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The membrane-bound HIV-1 envelope glycoprotein trimer precursor
| Entire | Name: The membrane-bound HIV-1 envelope glycoprotein trimer precursor |
|---|---|
| Components |
|
-Supramolecule #1000: The membrane-bound HIV-1 envelope glycoprotein trimer precursor
| Supramolecule | Name: The membrane-bound HIV-1 envelope glycoprotein trimer precursor type: sample / ID: 1000 Details: The membrane glycoprotein was monodisperse and solubilized in detergents. Oligomeric state: trimer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 440 KDa / Theoretical: 435 KDa / Method: Sedimentation, SDS-PAGE |
-Macromolecule #1: HIV-1 envelope glycoprotein trimer precursor
| Macromolecule | Name: HIV-1 envelope glycoprotein trimer precursor / type: protein_or_peptide / ID: 1 / Name.synonym: Env trimer, gp160 trimer Details: The membrane glycoprotein is solubilized in detergents. Number of copies: 3 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: JF-FL / synonym: HIV-1 / Location in cell: Plasma membrane Human immunodeficiency virus 1 / Strain: JF-FL / synonym: HIV-1 / Location in cell: Plasma membrane |
| Molecular weight | Experimental: 440 KDa / Theoretical: 435 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant plasmid: pcDNA3.1 in 293F cell line Homo sapiens (human) / Recombinant plasmid: pcDNA3.1 in 293F cell line |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Tris-HCl, 300 mM NaCl, 0.01% Cymal-6 |
| Grid | Details: 200 or 400 mesh copper grid with thin holey carbon support, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 93 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 89 K / Max: 93 K / Average: 91 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 150,000 times magnification |
| Date | Aug 1, 2009 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 200835 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder: CT3500 / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 10.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER, XIMPP / Number images used: 90306 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)