+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24952 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | tRNA-like Structure from Brome Mosaic Virus | ||||||||||||
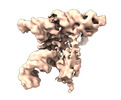
 Map data Map data | Cryo-EM map of brome mosaic virus (BMV) tRNA-like structure (TLS) generated using cryoSPARC. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Viral RNA / 3' UTR / RNA | ||||||||||||
| Biological species |   Brome mosaic virus Brome mosaic virus | ||||||||||||
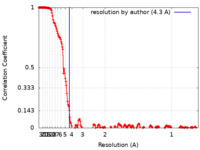
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | ||||||||||||
 Authors Authors | Kieft JS / Bonilla SL | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: A viral RNA hijacks host machinery using dynamic conformational changes of a tRNA-like structure. Authors: Steve L Bonilla / Madeline E Sherlock / Andrea MacFadden / Jeffrey S Kieft /  Abstract: Viruses require multifunctional structured RNAs to hijack their host’s biochemistry, but their mechanisms can be obscured by the difficulty of solving conformationally dynamic RNA structures. Using ...Viruses require multifunctional structured RNAs to hijack their host’s biochemistry, but their mechanisms can be obscured by the difficulty of solving conformationally dynamic RNA structures. Using cryo–electron microscopy (cryo-EM), we visualized the structure of the mysterious viral transfer RNA (tRNA)–like structure (TLS) from the brome mosaic virus, which affects replication, translation, and genome encapsidation. Structures in isolation and those bound to tyrosyl-tRNA synthetase (TyrRS) show that this ~55-kilodalton purported tRNA mimic undergoes large conformational rearrangements to bind TyrRS in a form that differs substantially from that of tRNA. Our study reveals how viral RNAs can use a combination of static and dynamic RNA structures to bind host machinery through highly noncanonical interactions, and we highlight the utility of cryo-EM for visualizing small, conformationally dynamic structured RNAs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24952.map.gz emd_24952.map.gz | 326.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24952-v30.xml emd-24952-v30.xml emd-24952.xml emd-24952.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24952_fsc.xml emd_24952_fsc.xml | 19.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_24952.png emd_24952.png | 85.4 KB | ||
| Filedesc metadata |  emd-24952.cif.gz emd-24952.cif.gz | 4.8 KB | ||
| Others |  emd_24952_additional_1.map.gz emd_24952_additional_1.map.gz emd_24952_half_map_1.map.gz emd_24952_half_map_1.map.gz emd_24952_half_map_2.map.gz emd_24952_half_map_2.map.gz | 630.4 MB 621 MB 621 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24952 http://ftp.pdbj.org/pub/emdb/structures/EMD-24952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24952 | HTTPS FTP |
-Related structure data
| Related structure data |  7samMC  7sc6C  7scqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10993 (Title: Cryo-EM micrographs of BMV TLS RNA / Data size: 6.7 TB EMPIAR-10993 (Title: Cryo-EM micrographs of BMV TLS RNA / Data size: 6.7 TBData #1: Aligned micrographs (with and without dose-weighting), dataset 1 [micrographs - single frame] Data #2: Aligned micrographs (with and without dose-weighting), dataset 2 [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24952.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24952.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of brome mosaic virus (BMV) tRNA-like structure (TLS) generated using cryoSPARC. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.413 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened (B factor: 257.7) cryo-EM map of brome...
| File | emd_24952_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened (B factor: 257.7) cryo-EM map of brome mosaic virus (BMV) tRNA-like structure (TLS) generated using cryoSPARC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A of brome mosaic virus (BMV) tRNA-like...
| File | emd_24952_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A of brome mosaic virus (BMV) tRNA-like structure (TLS) generated using cryoSPARC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B of brome mosaic virus (BMV) tRNA-like...
| File | emd_24952_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B of brome mosaic virus (BMV) tRNA-like structure (TLS) generated using cryoSPARC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : tRNA-like structure at the 3' UTR of the brome mosaic virus (BMV)...
| Entire | Name: tRNA-like structure at the 3' UTR of the brome mosaic virus (BMV) genome |
|---|---|
| Components |
|
-Supramolecule #1: tRNA-like structure at the 3' UTR of the brome mosaic virus (BMV)...
| Supramolecule | Name: tRNA-like structure at the 3' UTR of the brome mosaic virus (BMV) genome type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: RNA was generated by in vitro transcription of sequence-verified linear DNA template |
|---|---|
| Source (natural) | Organism:   Brome mosaic virus Brome mosaic virus |
| Molecular weight | Theoretical: 54.56 KDa |
-Macromolecule #1: Viral RNA
| Macromolecule | Name: Viral RNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Brome mosaic virus Brome mosaic virus |
| Molecular weight | Theoretical: 55.044559 KDa |
| Sequence | String: GGCGUGGUUG ACACGCAGAC CUCUUACAAG AGUGUCUAGG UGCCUUUGAG AGUUACUCUU UGCUCUCUUC GGAAGAACCC UUAGGGGUU CGUGCAUGGG CUUGCAUAGC AAGUCUUAGA AUGCGGGUAC CGUACAGUGU UGAAAAACAC UGUAAAUCUC U AAAAGAGA CCA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Concentration: 50.0 mM / Component - Name: NaMOPS / Details: Solution contained 10 mM magnesium chloride. |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse as determined by native gel electrophoresis. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -0.8 µm / Nominal defocus min: -2.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7sam: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)