[English] 日本語
 Yorodumi
Yorodumi- EMDB-22895: Low resolution map of the nucleosome remodelling and deacetylase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22895 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
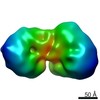
| Title | Low resolution map of the nucleosome remodelling and deacetylase complex from MEL cells. | |||||||||
 Map data Map data | Map of the nucleosome remodelling and deacetylase complex (NuRD) produced using negative-stained electron microscopy images analyzed in Cryosparc. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 14.0 Å | |||||||||
 Authors Authors | Jackman MJ / Landsberg MJ / Mackay JP | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020Title: The Nucleosome Remodeling and Deacetylase Complex Has an Asymmetric, Dynamic, and Modular Architecture. Authors: Jason K K Low / Ana P G Silva / Mehdi Sharifi Tabar / Mario Torrado / Sarah R Webb / Benjamin L Parker / Maryam Sana / Callum Smits / Jason W Schmidberger / Lou Brillault / Matthew J Jackman ...Authors: Jason K K Low / Ana P G Silva / Mehdi Sharifi Tabar / Mario Torrado / Sarah R Webb / Benjamin L Parker / Maryam Sana / Callum Smits / Jason W Schmidberger / Lou Brillault / Matthew J Jackman / David C Williams / Gerd A Blobel / Sandra B Hake / Nicholas E Shepherd / Michael J Landsberg / Joel P Mackay /    Abstract: The nucleosome remodeling and deacetylase (NuRD) complex is essential for metazoan development but has been refractory to biochemical analysis. We present an integrated analysis of the native ...The nucleosome remodeling and deacetylase (NuRD) complex is essential for metazoan development but has been refractory to biochemical analysis. We present an integrated analysis of the native mammalian NuRD complex, combining quantitative mass spectrometry, cross-linking, protein biochemistry, and electron microscopy to define the architecture of the complex. NuRD is built from a 2:2:4 (MTA, HDAC, and RBBP) deacetylase module and a 1:1:1 (MBD, GATAD2, and Chromodomain-Helicase-DNA-binding [CHD]) remodeling module, and the complex displays considerable structural dynamics. The enigmatic GATAD2 controls the asymmetry of the complex and directly recruits the CHD remodeler. The MTA-MBD interaction acts as a point of functional switching, with the transcriptional regulator PWWP2A competing with MBD for binding to the MTA-HDAC-RBBP subcomplex. Overall, our data address the long-running controversy over NuRD stoichiometry, provide imaging of the mammalian NuRD complex, and establish the biochemical mechanism by which PWWP2A can regulate NuRD composition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22895.map.gz emd_22895.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22895-v30.xml emd-22895-v30.xml emd-22895.xml emd-22895.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22895.png emd_22895.png | 67.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22895 http://ftp.pdbj.org/pub/emdb/structures/EMD-22895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22895 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10537 (Title: Negative-stained transmission electron microscopy of the nucleosome remodelling and deacetylase complex purified from murine erythroleukemia cells. EMPIAR-10537 (Title: Negative-stained transmission electron microscopy of the nucleosome remodelling and deacetylase complex purified from murine erythroleukemia cells.Data size: 82.7 Data #1: raw single-frame micrographs of the nucleosome remodelling and deacetylase complex (crosslinked). [micrographs - single frame])  EMPIAR-10538 (Title: Negative-stained transmission electron microscopy of the nucleosome and deacetylase complex purified from murine erythroleukemia cells. EMPIAR-10538 (Title: Negative-stained transmission electron microscopy of the nucleosome and deacetylase complex purified from murine erythroleukemia cells.Data size: 25.0 Data #1: raw single-frame micrographs of the nucleosome deacetylase complex (crosslinked). [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22895.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22895.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the nucleosome remodelling and deacetylase complex (NuRD) produced using negative-stained electron microscopy images analyzed in Cryosparc. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.17 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
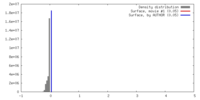
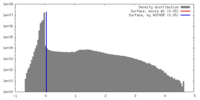
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Endogenous nucleosome remodelling and deacetylase complex purifie...
| Entire | Name: Endogenous nucleosome remodelling and deacetylase complex purified from MEL cells |
|---|---|
| Components |
|
-Supramolecule #1: Endogenous nucleosome remodelling and deacetylase complex purifie...
| Supramolecule | Name: Endogenous nucleosome remodelling and deacetylase complex purified from MEL cells type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Purified using a GST-tagged column bound by a recombinant GST-FOG1 protein; crosslinked using GraFix |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 955 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl acetate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 100.0 e/Å2 Details: Images recorded on a DE LC1100 lens coupled CCD detector. Estimated dose. |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)