+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21382 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM map of an MTA-HDAC-MBD complex | |||||||||
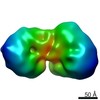
 Map data Map data | Negative stain EM map of an MTA-HDAC-MBD complex. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
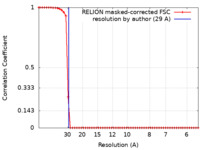
| Method | single particle reconstruction / negative staining / Resolution: 29.0 Å | |||||||||
 Authors Authors | Silva APG / Low JKK / Tabar MS / Torrado M / Schmidberger JW / Brillault L / Landsberg MJ / Mackay JP | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020Title: The Nucleosome Remodeling and Deacetylase Complex Has an Asymmetric, Dynamic, and Modular Architecture. Authors: Jason K K Low / Ana P G Silva / Mehdi Sharifi Tabar / Mario Torrado / Sarah R Webb / Benjamin L Parker / Maryam Sana / Callum Smits / Jason W Schmidberger / Lou Brillault / Matthew J Jackman ...Authors: Jason K K Low / Ana P G Silva / Mehdi Sharifi Tabar / Mario Torrado / Sarah R Webb / Benjamin L Parker / Maryam Sana / Callum Smits / Jason W Schmidberger / Lou Brillault / Matthew J Jackman / David C Williams / Gerd A Blobel / Sandra B Hake / Nicholas E Shepherd / Michael J Landsberg / Joel P Mackay /    Abstract: The nucleosome remodeling and deacetylase (NuRD) complex is essential for metazoan development but has been refractory to biochemical analysis. We present an integrated analysis of the native ...The nucleosome remodeling and deacetylase (NuRD) complex is essential for metazoan development but has been refractory to biochemical analysis. We present an integrated analysis of the native mammalian NuRD complex, combining quantitative mass spectrometry, cross-linking, protein biochemistry, and electron microscopy to define the architecture of the complex. NuRD is built from a 2:2:4 (MTA, HDAC, and RBBP) deacetylase module and a 1:1:1 (MBD, GATAD2, and Chromodomain-Helicase-DNA-binding [CHD]) remodeling module, and the complex displays considerable structural dynamics. The enigmatic GATAD2 controls the asymmetry of the complex and directly recruits the CHD remodeler. The MTA-MBD interaction acts as a point of functional switching, with the transcriptional regulator PWWP2A competing with MBD for binding to the MTA-HDAC-RBBP subcomplex. Overall, our data address the long-running controversy over NuRD stoichiometry, provide imaging of the mammalian NuRD complex, and establish the biochemical mechanism by which PWWP2A can regulate NuRD composition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21382.map.gz emd_21382.map.gz | 478.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21382-v30.xml emd-21382-v30.xml emd-21382.xml emd-21382.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21382_fsc.xml emd_21382_fsc.xml | 4.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_21382.png emd_21382.png | 20.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21382 http://ftp.pdbj.org/pub/emdb/structures/EMD-21382 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21382 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21382 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10539 (Title: Negative-stained transmission electron microscopy of the MTA-HDAC-MBD core complex purified from EXPI cells EMPIAR-10539 (Title: Negative-stained transmission electron microscopy of the MTA-HDAC-MBD core complex purified from EXPI cellsData size: 20.3 Data #1: micrograph of the MTA-HDAC-MBDcc core compelx of NuRD [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21382.map.gz / Format: CCP4 / Size: 12.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21382.map.gz / Format: CCP4 / Size: 12.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM map of an MTA-HDAC-MBD complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.79 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : MHm complex
| Entire | Name: MHm complex |
|---|---|
| Components |
|
-Supramolecule #1: MHm complex
| Supramolecule | Name: MHm complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Complex formed by HA-MTA2(1-429), HA-HDAC1 and FLAG-MBD3cc that were recombinantly expressed in HEK293 cells. |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
| Molecular weight | Experimental: 300 KDa |
-Supramolecule #2: Metastasis-associated protein MTA2,
| Supramolecule | Name: Metastasis-associated protein MTA2, / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
-Supramolecule #3: Histone deacetylase 1
| Supramolecule | Name: Histone deacetylase 1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Metastasis-associated protein MTA2
| Macromolecule | Name: Metastasis-associated protein MTA2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAANMYRVGD YVYFENSSSN PYLVRRIEEL NKTANGNVEA KVVCLFRRRD ISSSLNSLAD SNAREFEEE SKQPGVSEQQ RHQLKHRELF LSRQFESLPA THIRGKCSVT LLNETDILNQ Y LDKEDCFF YSLVFDPVQK TLLADQGEIR VGCKFQAEIP DRLAEGESDN ...String: MAANMYRVGD YVYFENSSSN PYLVRRIEEL NKTANGNVEA KVVCLFRRRD ISSSLNSLAD SNAREFEEE SKQPGVSEQQ RHQLKHRELF LSRQFESLPA THIRGKCSVT LLNETDILNQ Y LDKEDCFF YSLVFDPVQK TLLADQGEIR VGCKFQAEIP DRLAEGESDN RNQQKMEMKV WD PDNPLTD RQIDQFLVVA RAVGTFARAL DCSSSIRQPS LHMSAAAASR DITLFHAMDT LQR NGYDLA KAMSTLVPQG GPVLCRDEME EWSASEAMLF EEALEKYGKD FNDIRQDFLP WKSL ASIVQ FYYMWKTTDR YIQQKRLKAA EADSKLKQVY IPTYTKPNPN QIISVGSKPG MNGAG FQKG LTCESCHTTQ SAQWYAWGPP NMQCRLCASC WIYWKKYGGL KTPTQLEGAA RGTTEP HSR GHLSRP |
-Macromolecule #2: Histone deacetylase 1
| Macromolecule | Name: Histone deacetylase 1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAQTQGTRRK VCYYYDGDVG NYYYGQGHPM KPHRIRMTHN LLLNYGLYRK MEIYRPHKAN AEEMTKYHS DDYIKFLRSI RPDNMSEYSK QMQRFNVGED CPVFDGLFEF CQLSTGGSVA S AVKLNKQQ TDIAVNWAGG LHHAKKSEAS GFCYVNDIVL AILELLKYHQ ...String: MAQTQGTRRK VCYYYDGDVG NYYYGQGHPM KPHRIRMTHN LLLNYGLYRK MEIYRPHKAN AEEMTKYHS DDYIKFLRSI RPDNMSEYSK QMQRFNVGED CPVFDGLFEF CQLSTGGSVA S AVKLNKQQ TDIAVNWAGG LHHAKKSEAS GFCYVNDIVL AILELLKYHQ RVLYIDIDIH HG DGVEEAF YTTDRVMTVS FHKYGEYFPG TGDLRDIGAG KGKYYAVNYP LRDGIDDESY EAI FKPVMS KVMEMFQPSA VVLQCGSDSL SGDRLGCFNL TIKGHAKCVE FVKSFNLPML MLGG GGYTI RNVARCWTYE TAVALDTEIP NELPYNDYFE YFGPDFKLHI SPSNMTNQNT NEYLE KIKQ RLFENLRMLP HAPGVQMQAI PEDAIPEESG DEDEDDPDKR ISICSSDKRI ACEEEF SDS EEEGEGGRKN SSNFKKAKRV KTEDEKEKDP EEKKEVTEEE KTKEEKPEAK GVKEEVK LA |
-Macromolecule #3: Methyl-CpG-binding protein MBD3/GATA zinc finger domain-containin...
| Macromolecule | Name: Methyl-CpG-binding protein MBD3/GATA zinc finger domain-containing protein 2A fusion type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MERKRWECPA LPQGWEREEV PRRSGLSAGH RDVFYYSPSG KKFRSKPQLA RYLGGSMDLS TFDFRTGKM LMNKMNKSRQ RVRYDSSNQV KGKPDLNTAL PVRQTASIFK QPVTKITNHP S NKVKSDPQ KAVDQPRQLF WEKKLSGLSA FDIAEELVRT MDLPKGLQGV ...String: MERKRWECPA LPQGWEREEV PRRSGLSAGH RDVFYYSPSG KKFRSKPQLA RYLGGSMDLS TFDFRTGKM LMNKMNKSRQ RVRYDSSNQV KGKPDLNTAL PVRQTASIFK QPVTKITNHP S NKVKSDPQ KAVDQPRQLF WEKKLSGLSA FDIAEELVRT MDLPKGLQGV GPGCTDETLL SA IASALHT STLPITGQLS AAVEKNPGVW LNTAQPLCKA FMVTDDDIRK QEELVQQVRK RLE EALMAD MLAHVEELAR DGEAPLDKAC AEEEEEEEEE EEEPEPERVS PEERERMIKQ LKEELRLEEA KLVLLKKLRQ SQIQKEATAQ K |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.027 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: Sample (5 microliters) was applied to a glow-discharged, carbon-coated 400-mesh copper grid (GSCu400CC, ProSciTech). After 2 minutes incubation time, the grid was blotted and washed with ten ...Details: Sample (5 microliters) was applied to a glow-discharged, carbon-coated 400-mesh copper grid (GSCu400CC, ProSciTech). After 2 minutes incubation time, the grid was blotted and washed with ten drops of distilled water, blotting on a filter paper in between washes. The grid was then washed in a drop of 1 % (w/v) uranyl acetate, blotted and subsequently incubated in another drop of 1% (w/v) uranyl acetate solution for 30 seconds, blotted and allowed to air dry at room temperature. | ||||||||||||
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Details | Sample was a sample from GraFix purification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: OTHER / Number grids imaged: 1 / Number real images: 325 / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)