[English] 日本語
 Yorodumi
Yorodumi- EMDB-14722: Cryo-EM structure of USP1-UAF1 bound to FANCI and mono-ubiquitina... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of USP1-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with ML323 (consensus reconstruction) | |||||||||
 Map data Map data | Globally sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Deubiquitinase / Complex / Enzyme-Substrate / Inhibitor / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of protein monoubiquitination / positive regulation of error-prone translesion synthesis / Signaling by cytosolic PDGFRA and PDGFRB fusion proteins / regulation of CD40 signaling pathway / gamete generation / monoubiquitinated protein deubiquitination / regulation of regulatory T cell differentiation / double-strand break repair involved in meiotic recombination / homologous chromosome pairing at meiosis / neuronal stem cell population maintenance ...regulation of protein monoubiquitination / positive regulation of error-prone translesion synthesis / Signaling by cytosolic PDGFRA and PDGFRB fusion proteins / regulation of CD40 signaling pathway / gamete generation / monoubiquitinated protein deubiquitination / regulation of regulatory T cell differentiation / double-strand break repair involved in meiotic recombination / homologous chromosome pairing at meiosis / neuronal stem cell population maintenance / brain morphogenesis / deubiquitinase activator activity / DNA repair complex / mitotic intra-S DNA damage checkpoint signaling / skeletal system morphogenesis / skin development / seminiferous tubule development / homeostasis of number of cells / protein deubiquitination / embryonic organ development / single fertilization / interstrand cross-link repair / regulation of DNA repair / response to UV / positive regulation of double-strand break repair via homologous recombination / condensed chromosome / DNA polymerase binding / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / Regulation of pyruvate metabolism / NF-kB is activated and signals survival / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Downregulation of ERBB2:ERBB3 signaling / Regulation of innate immune responses to cytosolic DNA / Pexophagy / NRIF signals cell death from the nucleus / Activated NOTCH1 Transmits Signal to the Nucleus / Regulation of PTEN localization / VLDLR internalisation and degradation / ubiquitin binding / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / positive regulation of protein ubiquitination / TICAM1, RIP1-mediated IKK complex recruitment / Regulation of BACH1 activity / Translesion synthesis by REV1 / MAP3K8 (TPL2)-dependent MAPK1/3 activation / Degradation of CDH1 / Translesion synthesis by POLK / InlB-mediated entry of Listeria monocytogenes into host cell / positive regulation of epithelial cell proliferation / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / Josephin domain DUBs / Downregulation of TGF-beta receptor signaling / Translesion synthesis by POLI / response to gamma radiation / Gap-filling DNA repair synthesis and ligation in GG-NER / IKK complex recruitment mediated by RIP1 / Degradation of CRY and PER proteins / Regulation of activated PAK-2p34 by proteasome mediated degradation / PINK1-PRKN Mediated Mitophagy / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / TNFR1-induced NF-kappa-B signaling pathway / skeletal system development / Autodegradation of Cdh1 by Cdh1:APC/C / TCF dependent signaling in response to WNT / Regulation of NF-kappa B signaling / APC/C:Cdc20 mediated degradation of Securin / positive regulation of receptor signaling pathway via JAK-STAT / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / activated TAK1 mediates p38 MAPK activation / TP53 Regulates Transcription of DNA Repair Genes / Asymmetric localization of PCP proteins Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | |||||||||
 Authors Authors | Rennie ML / Walden H | |||||||||
| Funding support | European Union,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site. Authors: Martin L Rennie / Connor Arkinson / Viduth K Chaugule / Helen Walden /  Abstract: Repair of DNA damage is critical to genomic integrity and frequently disrupted in cancers. Ubiquitin-specific protease 1 (USP1), a nucleus-localized deubiquitinase, lies at the interface of multiple ...Repair of DNA damage is critical to genomic integrity and frequently disrupted in cancers. Ubiquitin-specific protease 1 (USP1), a nucleus-localized deubiquitinase, lies at the interface of multiple DNA repair pathways and is a promising drug target for certain cancers. Although multiple inhibitors of this enzyme, including one in phase 1 clinical trials, have been established, their binding mode is unknown. Here, we use cryo-electron microscopy to study an assembled enzyme-substrate-inhibitor complex of USP1 and the well-established inhibitor, ML323. Achieving 2.5-Å resolution, with and without ML323, we find an unusual binding mode in which the inhibitor disrupts part of the hydrophobic core of USP1. The consequent conformational changes in the secondary structure lead to subtle rearrangements in the active site that underlie the mechanism of inhibition. These structures provide a platform for structure-based drug design targeting USP1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14722.map.gz emd_14722.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14722-v30.xml emd-14722-v30.xml emd-14722.xml emd-14722.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14722_fsc.xml emd_14722_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14722.png emd_14722.png | 179.7 KB | ||
| Masks |  emd_14722_msk_1.map emd_14722_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14722.cif.gz emd-14722.cif.gz | 8.1 KB | ||
| Others |  emd_14722_half_map_1.map.gz emd_14722_half_map_1.map.gz emd_14722_half_map_2.map.gz emd_14722_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14722 http://ftp.pdbj.org/pub/emdb/structures/EMD-14722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14722 | HTTPS FTP |
-Related structure data
| Related structure data |  8a9kMC  7zh3C  7zh4C  8a9jC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14722.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14722.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Globally sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
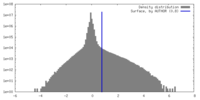
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14722_msk_1.map emd_14722_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
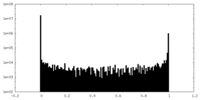
| Density Histograms |
-Half map: #1
| File | emd_14722_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
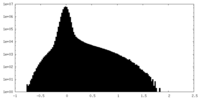
| Density Histograms |
-Half map: #2
| File | emd_14722_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : USP1(C90S)-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with...
| Entire | Name: USP1(C90S)-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with dsDNA and ML323 |
|---|---|
| Components |
|
-Supramolecule #1: USP1(C90S)-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with...
| Supramolecule | Name: USP1(C90S)-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with dsDNA and ML323 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Fanconi anemia group I protein, Fanconi anemia group D2 protein w...
| Supramolecule | Name: Fanconi anemia group I protein, Fanconi anemia group D2 protein with ubiquitin conjugated to K561, Ubiquitin carboxyl-terminal hydrolase 1, WD repeat-containing protein 48 type: complex / ID: 2 / Parent: 1 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fanconi anemia group I protein
| Macromolecule | Name: Fanconi anemia group I protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHMDQ KILSLAAEKT ADKLQEFLQT LREGDLTNLL QNQAVKGKVA GALLRAIFKG SPCSEEAGTL RRRKIYTCCI QLVESGDLQK EIASEIIGLL MLEAHHFPGP LLVELANEFI SAVREGSLVN GKSLELLPII LTALATKKEN LAYGKGVLSG EECKKQLINT ...String: MHHHHHHMDQ KILSLAAEKT ADKLQEFLQT LREGDLTNLL QNQAVKGKVA GALLRAIFKG SPCSEEAGTL RRRKIYTCCI QLVESGDLQK EIASEIIGLL MLEAHHFPGP LLVELANEFI SAVREGSLVN GKSLELLPII LTALATKKEN LAYGKGVLSG EECKKQLINT LCSGRWDQQY VIQLTSMFKD VPLTAEEVEF VVEKALSMFS KMNLQEIPPL VYQLLVLSSK GSRKSVLEGI IAFFSALDKQ HNEEQSGDEL LDVVTVPSGE LRHVEGTIIL HIVFAIKLDY ELGRELVKHL KVGQQGDSNN NLSPFSIALL LSVTRIQRFQ DQVLDLLKTS VVKSFKDLQL LQGSKFLQNL VPHRSYVSTM ILEVVKNSVH SWDHVTQGLV ELGFILMDSY GPKKVLDGKT IETSPSLSRM PNQHACKLGA NILLETFKIH EMIRQEILEQ VLNRVVTRAS SPISHFLDLL SNIVMYAPLV LQSCSSKVTE AFDYLSFLPL QTVQRLLKAV QPLLKVSMSM RDCLILVLRK AMFANQLDAR KSAVAGFLLL LKNFKVLGSL SSSQCSQSLS VSQVHVDVHS HYNSVANETF CLEIMDSLRR CLSQQADVRL MLYEGFYDVL RRNSQLANSV MQTLLSQLKQ FYEPKPDLLP PLKLEACILT QGDKISLQEP LDYLLCCIQH CLAWYKNTVI PLQQGEEEEE EEEAFYEDLD DILESITNRM IKSELEDFEL DKSADFSQST SIGIKNNICA FLVMGVCEVL IEYNFSISSF SKNRFEDILS LFMCYKKLSD ILNEKAGKAK TKMANKTSDS LLSMKFVSSL LTALFRDSIQ SHQESLSVLR SSNEFMRYAV NVALQKVQQL KETGHVSGPD GQNPEKIFQN LCDITRVLLW RYTSIPTSVE ESGKKEKGKS ISLLCLEGLQ KIFSAVQQFY QPKIQQFLRA LDVTDKEGEE REDADVSVTQ RTAFQIRQFQ RSLLNLLSSQ EEDFNSKEAL LLVTVLTSLS KLLEPSSPQF VQMLSWTSKI CKENSREDAL FCKSLMNLLF SLHVSYKSPV ILLRDLSQDI HGHLGDIDQD VEVEKTNHFA IVNLRTAAPT VCLLVLSQAE KVLEEVDWLI TKLKGQVSQE TLSEEASSQA TLPNQPVEKA IIMQLGTLLT FFHELVQTAL PSGSCVDTLL KDLCKMYTTL TALVRYYLQV CQSSGGIPKN MEKLVKLSGS HLTPLCYSFI SYVQNKSKSL NYTGEKKEKP AAVATAMARV LRETKPIPNL IFAIEQYEKF LIHLSKKSKV NLMQHMKLST SRDFKIKGNI LDMVLREDGE DENEEGTASE HGGQNKEPAK KKRKK UniProtKB: Fanconi anemia group I protein |
-Macromolecule #2: Fanconi anemia group D2 protein
| Macromolecule | Name: Fanconi anemia group D2 protein / type: protein_or_peptide / ID: 2 / Details: Ubiquitin conjugated to K561 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMVSKRR LSKSEDKESL TEDASKTRKQ PLSKKTKKSH IANEVEENDS IFVKLLKISG IILKTGESQN QLAVDQIAFQ KKLFQTLRRH PSYPKIIEEF VSGLESYIED EDSFRNCLLS CERLQDEEAS MGASYSKSLI KLLLGIDILQ PAIIKTLFEK LPEYFFENKN ...String: GPGSMVSKRR LSKSEDKESL TEDASKTRKQ PLSKKTKKSH IANEVEENDS IFVKLLKISG IILKTGESQN QLAVDQIAFQ KKLFQTLRRH PSYPKIIEEF VSGLESYIED EDSFRNCLLS CERLQDEEAS MGASYSKSLI KLLLGIDILQ PAIIKTLFEK LPEYFFENKN SDEINIPRLI VSQLKWLDRV VDGKDLTTKI MQLISIAPEN LQHDIITSLP EILGDSQHAD VGKELSDLLI ENTSLTVPIL DVLSSLRLDP NFLLKVRQLV MDKLSSIRLE DLPVIIKFIL HSVTAMDTLE VISELREKLD LQHCVLPSRL QASQVKLKSK GRASSSGNQE SSGQSCIILL FDVIKSAIRY EKTISEAWIK AIENTASVSE HKVFDLVMLF IIYSTNTQTK KYIDRVLRNK IRSGCIQEQL LQSTFSVHYL VLKDMCSSIL SLAQSLLHSL DQSIISFGSL LYKYAFKFFD TYCQQEVVGA LVTHICSGNE AEVDTALDVL LELVVLNPSA MMMNAVFVKG ILDYLDNISP QQIRKLFYVL STLAFSKQNE ASSHIQDDMH LVIRKQLSST VFKYKLIGII GAVTMAGIMA ADRSESPSLT QERANLSDEQ CTQVTSLLQL VHSCSEQSPQ ASALYYDEFA NLIQHEKLDP KALEWVGHTI CNDFQDAFVV DSCVVPEGDF PFPVKALYGL EEYDTQDGIA INLLPLLFSQ DFAKDGGPVT SQESGQKLVS PLCLAPYFRL LRLCVERQHN GNLEEIDGLL DCPIFLTDLE PGEKLESMSA KERSFMCSLI FLTLNWFREI VNAFCQETSP EMKGKVLTRL KHIVELQIIL EKYLAVTPDY VPPLGNFDVE TLDITPHTVT AISAKIRKKG KIERKQKTDG SKTSSSDTLS EEKNSECDPT PSHRGQLNKE FTGKEEKTSL LLHNSHAFFR ELDIEVFSIL HCGLVTKFIL DTEMHTEATE VVQLGPPELL FLLEDLSQKL ESMLTPPIAR RVPFLKNKGS RNIGFSHLQQ RSAQEIVHCV FQLLTPMCNH LENIHNYFQC LAAENHGVVD GPGVKVQEYH IMSSCYQRLL QIFHGLFAWS GFSQPENQNL LYSALHVLSS RLKQGEHSQP LEELLSQSVH YLQNFHQSIP SFQCALYLIR LLMVILEKST ASAQNKEKIA SLARQFLCRV WPSGDKEKSN ISNDQLHALL CIYLEHTESI LKAIEEIAGV GVPELINSPK DASSSTFPTL TRHTFVVFFR VMMAELEKTV KKIEPGTAAD SQQIHEEKLL YWNMAVRDFS ILINLIKVFD SHPVLHVCLK YGRLFVEAFL KQCMPLLDFS FRKHREDVLS LLETFQLDTR LLHHLCGHSK IHQDTRLTQH VPLLKKTLEL LVCRVKAMLT LNNCREAFWL GNLKNRDLQG EEIKSQNSQE STADESEDDM SSQASKSKAT EDGEEDEVSA GEKEQDSDES YDDSD UniProtKB: Fanconi anemia group D2 protein |
-Macromolecule #3: Polyubiquitin-C
| Macromolecule | Name: Polyubiquitin-C / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMQIFVK TLTGKTITLE VEPSDTIENV KAKIQDKEGI PPDQQRLIFA GKQLEDGRTL SDYNIQKEST LHLVLRLRGG |
-Macromolecule #4: Ubiquitin carboxyl-terminal hydrolase 1
| Macromolecule | Name: Ubiquitin carboxyl-terminal hydrolase 1 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GMPGVIPSES NGLSRGSPSK KNRLSLKFFQ KKETKRALDF TDSQENEEKA SEYRASEIDQ VVPAAQSSPI NCEKRENLLP FVGLNNLGNT SYLNSILQVL YFCPGFKSGV KHLFNIISRK KEALKDEANQ KDKGNCKEDS LASYELICSL QSLIISVEQL QASFLLNPEK ...String: GMPGVIPSES NGLSRGSPSK KNRLSLKFFQ KKETKRALDF TDSQENEEKA SEYRASEIDQ VVPAAQSSPI NCEKRENLLP FVGLNNLGNT SYLNSILQVL YFCPGFKSGV KHLFNIISRK KEALKDEANQ KDKGNCKEDS LASYELICSL QSLIISVEQL QASFLLNPEK YTDELATQPR RLLNTLRELN PMYEGYLQHD AQEVLQCILG NIQETCQLLK KEEVKNVAEL PTKVEEIPHP KEEMNGINSI EMDSMRHSED FKEKLPKGNG KRKSDTEFGN MKKKVKLSKE HQSLEENQRQ TRSKRKATSD TLESPPKIIP KYISENESPR PSQKKSRVKI NWLKSATKQP SILSKFCSLG KITTNQGVKG QSKENECDPE EDLGKCESDN TTNGCGLESP GNTVTPVNVN EVKPINKGEE QIGFELVEKL FQGQLVLRTR CLECESLTER REDFQDISVP VQEDELSKVE ESSEISPEPK TEMKTLRWAI SQFASVERIV GEDKYFCENC HHYTEAERSL LFDKMPEVIT IHLKCFAASG LEFDCYGGGL SKINTPLLTP LKLSLEEWST KPTNDSYGLF AVVMHSGITI SSGHYTASVK VTDLNSLELD KGNFVVDQMC EIGKPEPLNE EEARGVVENY NDEEVSIRVG GNTQPSKVLN KKNVEAIGLL GGQKSKADYE LYNKASNPDK VASTAFAENR NSETSDTTGT HESDRNKESS DQTGINISGF ENKISYVVQS LKEYEGKWLL FDDSEVKVTE EKDFLNSLSP STSPTSTPYL LFYKKL UniProtKB: Ubiquitin carboxyl-terminal hydrolase 1 |
-Macromolecule #5: WD repeat-containing protein 48
| Macromolecule | Name: WD repeat-containing protein 48 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSMAA HHRQNTAGRR KVQVSYVIRD EVEKYNRNGV NALQLDPALN RLFTAGRDSI IRIWSVNQHK QDPYIASMEH HTDWVNDIVL CCNGKTLISA SSDTTVKVWN AHKGFCMSTL RTHKDYVKAL AYAKDKELVA SAGLDRQIFL WDVNTLTALT ...String: MHHHHHHLEV LFQGPGSMAA HHRQNTAGRR KVQVSYVIRD EVEKYNRNGV NALQLDPALN RLFTAGRDSI IRIWSVNQHK QDPYIASMEH HTDWVNDIVL CCNGKTLISA SSDTTVKVWN AHKGFCMSTL RTHKDYVKAL AYAKDKELVA SAGLDRQIFL WDVNTLTALT ASNNTVTTSS LSGNKDSIYS LAMNQLGTII VSGSTEKVLR VWDPRTCAKL MKLKGHTDNV KALLLNRDGT QCLSGSSDGT IRLWSLGQQR CIATYRVHDE GVWALQVNDA FTHVYSGGRD RKIYCTDLRN PDIRVLICEE KAPVLKMELD RSADPPPAIW VATTKSTVNK WTLKGIHNFR ASGDYDNDCT NPITPLCTQP DQVIKGGASI IQCHILNDKR HILTKDTNNN VAYWDVLKAC KVEDLGKVDF EDEIKKRFKM VYVPNWFSVD LKTGMLTITL DESDCFAAWV SAKDAGFSSP DGSDPKLNLG GLLLQALLEY WPRTHVNPMD EEENEVNHVN GEQENRVQKG NGYFQVPPHT PVIFGEAGGR TLFRLLCRDS GGETESMLLN ETVPQWVIDI TVDKNMPKFN KIPFYLQPHA SSGAKTLKKD RLSASDMLQV RKVMEHVYEK IINLDNESQT TSSSNNEKPG EQEKEEDIAV LAEEKIELLC QDQVLDPNMD LRTVKHFIWK SGGDLTLHYR QKST UniProtKB: WD repeat-containing protein 48 |
-Macromolecule #6: DNA (61-MER)
| Macromolecule | Name: DNA (61-MER) / type: dna / ID: 6 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: TGATCAGAGG TCATTTGAAT TCATGGCTTC GAGCTTCATG TAGAGTCGAC GGTGCTGGGA T |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 288 K / Details: blotted for 3.0 secs before plunging. |
| Details | 9.3 uM USP1-UAF1, 1.8 uM FANCI-FANCD2Ub, 2.2 uM dsDNA (61 base-pairs), 18 uM ML323 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Overall B value: 95.3 |
|---|---|
| Output model |  PDB-8a9k: |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)