[English] 日本語
 Yorodumi
Yorodumi- EMDB-11630: Envelope glycprotein of endogenous retrovirus Y032 (Atlas virus) ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11630 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Envelope glycprotein of endogenous retrovirus Y032 (Atlas virus) from the human hookworm Ancylostoma ceylanicum | ||||||||||||
 Map data Map data | Final EM map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | class II membrane fusion protein / retroviral envelope protein (Env) / lipid binding protein / disulfide bonding / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Ancylostoma ceylanicum (invertebrata) Ancylostoma ceylanicum (invertebrata) | ||||||||||||
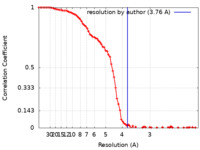
| Method | single particle reconstruction / cryo EM / Resolution: 3.76 Å | ||||||||||||
 Authors Authors | Mata CP / Merchant M | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: A bioactive phlebovirus-like envelope protein in a hookworm endogenous virus. Authors: Monique Merchant / Carlos P Mata / Yangci Liu / Haoming Zhai / Anna V Protasio / Yorgo Modis /  Abstract: Endogenous viral elements (EVEs), accounting for 15% of our genome, serve as a genetic reservoir from which new genes can emerge. Nematode EVEs are particularly diverse and informative of virus ...Endogenous viral elements (EVEs), accounting for 15% of our genome, serve as a genetic reservoir from which new genes can emerge. Nematode EVEs are particularly diverse and informative of virus evolution. We identify Atlas virus-an intact retrovirus-like EVE in the human hookworm , with an envelope protein genetically related to G-G glycoproteins from the family Phenuiviridae. A cryo-EM structure of Atlas G reveals a class II viral membrane fusion protein fold not previously seen in retroviruses. Atlas G has the structural hallmarks of an active fusogen. Atlas G trimers insert into membranes with endosomal lipid compositions and low pH. When expressed on the plasma membrane, Atlas G has cell-cell fusion activity. With its preserved biological activities, Atlas G has the potential to acquire a cellular function. Our work reveals structural plasticity in reverse-transcribing RNA viruses. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: A bioactive phlebovirus-like envelope protein in a hookworm endogenous virus Authors: Merchant M / Mata CP / Liu Y / Zhai H / Protasio AV / Modis Y | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11630.map.gz emd_11630.map.gz | 39.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11630-v30.xml emd-11630-v30.xml emd-11630.xml emd-11630.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11630_fsc.xml emd_11630_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_11630.png emd_11630.png | 167.6 KB | ||
| Masks |  emd_11630_msk_1.map emd_11630_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11630.cif.gz emd-11630.cif.gz | 7.4 KB | ||
| Others |  emd_11630_half_map_1.map.gz emd_11630_half_map_1.map.gz emd_11630_half_map_2.map.gz emd_11630_half_map_2.map.gz | 33 MB 33 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11630 http://ftp.pdbj.org/pub/emdb/structures/EMD-11630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11630 | HTTPS FTP |
-Related structure data
| Related structure data |  7a4aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10266 (Title: CryoEM image reconstuction of the envelope protein of endogenous retrovirus Y032 from the human hookworm Ancylostoma ceylanicum EMPIAR-10266 (Title: CryoEM image reconstuction of the envelope protein of endogenous retrovirus Y032 from the human hookworm Ancylostoma ceylanicumData size: 5.6 TB Data #1: Unaligned multi-frame micrographs of Env protein endogenous retrovirus AceY032 from Ancylostoma ceylanicum [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11630.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11630.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final EM map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11630_msk_1.map emd_11630_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_11630_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_11630_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Viral envelope glycoprotein
| Entire | Name: Viral envelope glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Viral envelope glycoprotein
| Supramolecule | Name: Viral envelope glycoprotein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Envelope glycoprotein of endogenous retrovirus Y032 (Atlas virus) from Ancylostoma ceylanicum |
|---|---|
| Source (natural) | Organism:  Ancylostoma ceylanicum (invertebrata) Ancylostoma ceylanicum (invertebrata) |
| Molecular weight | Theoretical: 143 KDa |
-Macromolecule #1: Integrase catalytic domain-containing protein
| Macromolecule | Name: Integrase catalytic domain-containing protein / type: protein_or_peptide / ID: 1 Details: In chains A, B and C, residue Asn414 is covalently modified with an N-linked N-acetyl glucosamine ligand. Chains A, B and C each contain the following 15 disulfide bonds: Cys1-Cys41, Cys14- ...Details: In chains A, B and C, residue Asn414 is covalently modified with an N-linked N-acetyl glucosamine ligand. Chains A, B and C each contain the following 15 disulfide bonds: Cys1-Cys41, Cys14-Cys23, Cys66-Cys162, Cys87-Cys135, Cys93-Cys142, Cys98-Cys123, Cys127-Cys132, Cys129-Cys138, Cys246-Cys257, Cys264-Cys277, Cys266-Cys275, Cys337-Cys408, Cys347-Cys350, Cys360-Cys382, Cys373-Cys404. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ancylostoma ceylanicum (invertebrata) Ancylostoma ceylanicum (invertebrata) |
| Molecular weight | Theoretical: 51.51909 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSEVISVTST EEVCTIQENK ETCTFNHATT ITLQPLQQQT CLTLNDPEKR PMGMLTVKPD GIKFRCNKKI EFFTRDHQIV SESVHRCHR AGSCHSDECH HVKDTDALPE FSSEANSRPG YTSCSSSCGC ITCDGCFFCE PSCLFHRLYA IPTTPTIYSI F YCPSWELE ...String: CSEVISVTST EEVCTIQENK ETCTFNHATT ITLQPLQQQT CLTLNDPEKR PMGMLTVKPD GIKFRCNKKI EFFTRDHQIV SESVHRCHR AGSCHSDECH HVKDTDALPE FSSEANSRPG YTSCSSSCGC ITCDGCFFCE PSCLFHRLYA IPTTPTIYSI F YCPSWELE VDAEISLQRE DETTTSTIRL LPGRTSTWNN IRFSLIGTIV PQLPILSSAF VTNGRQTSIV KPAYAGQLQS NS VGQLQCP NLEAAKQFEC HFSRNLCTCT NALHKVSCTC YDGSVEDHME ALPLPQTSKN FLVFEKDRNI YAKTHVGSAL QLH IVAQDL KITTVKHTSH CQVEASDLSG CYSCTSGASL TLSCKSDNGE VLANMKCNEQ THVIRCTESG FINNILLMFD TSEV AADCT AACPGGIVNF TIKGLLAFVN ERIISQSYSA TDVERNIKGK PIPNPLLGLD STRTGHHHHH H UniProtKB: Integrase catalytic domain-containing protein |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20 mM Tris/HCl (NH12C4O3Cl) 0.1 M NaCl 'sodium chloride' 5 % glycerol (C3H8O3) 0.5 mM TCEP (C9H15O6P) | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.2 K / Instrument: FEI VITROBOT MARK IV / Details: Grids were blotted for 4 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: OTHER / Spherical aberration corrector: None / Chromatic aberration corrector: None / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 3027 / Average exposure time: 8.0 sec. / Average electron dose: 46.18 e/Å2 / Details: Dose rate = 1.28 e- A^-2 per frame |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 75000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.3 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 691-1136 / Chain - Source name: PDB / Chain - Initial model type: experimental model Details: A homology model was built from PDB:6EGU using the Swiss-Model server (swissmodel.expasy.org). The model was docked as a rigid body into the density with UCSF Chimera prior to refinement. |
|---|---|
| Details | A homology model was built from PDB:6EGU using the Swiss-Model server (swissmodel.expasy.org). The model was docked as a rigid body into the density with UCSF Chimera prior to refinement. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 82 / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-7a4a: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)