[English] 日本語
 Yorodumi
Yorodumi- EMDB-10641: Structure of ryanodine receptor 1 in native membrane in the prese... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10641 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
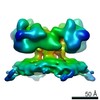
| Title | Structure of ryanodine receptor 1 in native membrane in the presence of DDM and EDTA | |||||||||
 Map data Map data | Structure of rabbit ryanodine receptor 1 in native membrane in the presence of DDM and EDTA | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 25.7 Å | |||||||||
 Authors Authors | Chen W / Kudryashev M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2020 Journal: EMBO Rep / Year: 2020Title: Structure of RyR1 in native membranes. Authors: Wenbo Chen / Mikhail Kudryashev /  Abstract: Ryanodine receptor 1 (RyR1) mediates excitation-contraction coupling by releasing Ca from sarcoplasmic reticulum (SR) to the cytoplasm of skeletal muscle cells. RyR1 activation is regulated by ...Ryanodine receptor 1 (RyR1) mediates excitation-contraction coupling by releasing Ca from sarcoplasmic reticulum (SR) to the cytoplasm of skeletal muscle cells. RyR1 activation is regulated by several proteins from both the cytoplasm and lumen of the SR. Here, we report the structure of RyR1 from native SR membranes in closed and open states. Compared to the previously reported structures of purified RyR1, our structure reveals helix-like densities traversing the bilayer approximately 5 nm from the RyR1 transmembrane domain and sarcoplasmic extensions linking RyR1 to a putative calsequestrin network. We document the primary conformation of RyR1 in situ and its structural variations. The activation of RyR1 is associated with changes in membrane curvature and movement in the sarcoplasmic extensions. Our results provide structural insight into the mechanism of RyR1 in its native environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
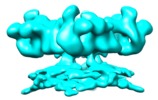
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10641.map.gz emd_10641.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10641-v30.xml emd-10641-v30.xml emd-10641.xml emd-10641.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10641.png emd_10641.png | 130.5 KB | ||
| Masks |  emd_10641_msk_1.map emd_10641_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Others |  emd_10641_half_map_1.map.gz emd_10641_half_map_1.map.gz emd_10641_half_map_2.map.gz emd_10641_half_map_2.map.gz | 3.6 MB 3.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10641 http://ftp.pdbj.org/pub/emdb/structures/EMD-10641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10641 | HTTPS FTP |
-Validation report
| Summary document |  emd_10641_validation.pdf.gz emd_10641_validation.pdf.gz | 357 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10641_full_validation.pdf.gz emd_10641_full_validation.pdf.gz | 356.1 KB | Display | |
| Data in XML |  emd_10641_validation.xml.gz emd_10641_validation.xml.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10641 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10641 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10349 (Title: Cryo Electron Tomograms of Membrane Fractions of Rabbit Skeletal Muscle for Structural Determination of RyR1 in SR Vesicles EMPIAR-10349 (Title: Cryo Electron Tomograms of Membrane Fractions of Rabbit Skeletal Muscle for Structural Determination of RyR1 in SR VesiclesData size: 254.2 Data #1: 82 tilt series (frames aligned my motioncorr2) in .st stacks (mrc format for imod) including .tlt, .xf and .defocus files [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10641.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10641.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of rabbit ryanodine receptor 1 in native membrane in the presence of DDM and EDTA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10641_msk_1.map emd_10641_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
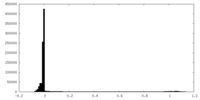
| Density Histograms |
-Half map: Structure of rabbit ryanodine receptor 1 in native...
| File | emd_10641_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of rabbit ryanodine receptor 1 in native membrane in the presence of DDM and EDTA (half map 2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
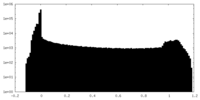
| Density Histograms |
-Half map: Structure of rabbit ryanodine receptor 1 in native...
| File | emd_10641_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of rabbit ryanodine receptor 1 in native membrane in the presence of DDM and EDTA (half map 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
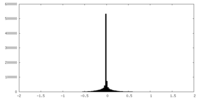
| Density Histograms |
- Sample components
Sample components
-Entire : Rabbit ryanodine receptor 1 in native membrane in the presence of...
| Entire | Name: Rabbit ryanodine receptor 1 in native membrane in the presence of DDM and EDTA |
|---|---|
| Components |
|
-Supramolecule #1: Rabbit ryanodine receptor 1 in native membrane in the presence of...
| Supramolecule | Name: Rabbit ryanodine receptor 1 in native membrane in the presence of DDM and EDTA type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7.1 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)