[English] 日本語
 Yorodumi
Yorodumi- EMDB-10430: Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10430 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related Human variant R4810K | |||||||||
 Map data Map data | Composite map assembled from 4 focused refinements | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNF213 / E3 LIgase / AAA-Protein / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlipid ubiquitination / negative regulation of non-canonical Wnt signaling pathway / lipid droplet formation / xenophagy / Antigen processing: Ubiquitination & Proteasome degradation / sprouting angiogenesis / Transferases; Acyltransferases; Aminoacyltransferases / regulation of lipid metabolic process / protein K63-linked ubiquitination / immune system process ...lipid ubiquitination / negative regulation of non-canonical Wnt signaling pathway / lipid droplet formation / xenophagy / Antigen processing: Ubiquitination & Proteasome degradation / sprouting angiogenesis / Transferases; Acyltransferases; Aminoacyltransferases / regulation of lipid metabolic process / protein K63-linked ubiquitination / immune system process / protein autoubiquitination / lipid droplet / RING-type E3 ubiquitin transferase / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / angiogenesis / ubiquitin-dependent protein catabolic process / defense response to bacterium / protein ubiquitination / ATP hydrolysis activity / zinc ion binding / ATP binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Ahel J / Meinhart A | |||||||||
| Funding support |  Austria, 1 items Austria, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism. Authors: Juraj Ahel / Anita Lehner / Antonia Vogel / Alexander Schleiffer / Anton Meinhart / David Haselbach / Tim Clausen /  Abstract: RNF213 is the major susceptibility factor for Moyamoya disease, a progressive cerebrovascular disorder that often leads to brain stroke in adults and children. Characterization of disease-associated ...RNF213 is the major susceptibility factor for Moyamoya disease, a progressive cerebrovascular disorder that often leads to brain stroke in adults and children. Characterization of disease-associated mutations has been complicated by the enormous size of RNF213. Here, we present the cryo-EM structure of mouse RNF213. The structure reveals the intricate fold of the 584 kDa protein, comprising an N-terminal stalk, a dynein-like core with six ATPase units, and a multidomain E3 module. Collaboration with UbcH7, a cysteine-reactive E2, points to an unexplored ubiquitin-transfer mechanism that proceeds in a RING-independent manner. Moreover, we show that pathologic MMD mutations cluster in the composite E3 domain, likely interfering with substrate ubiquitination. In conclusion, the structure of RNF213 uncovers a distinct type of an E3 enzyme, highlighting the growing mechanistic diversity in ubiquitination cascades. Our results also provide the molecular framework for investigating the emerging role of RNF213 in lipid metabolism, hypoxia, and angiogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10430.map.gz emd_10430.map.gz | 153.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10430-v30.xml emd-10430-v30.xml emd-10430.xml emd-10430.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10430.png emd_10430.png | 51 KB | ||
| Filedesc metadata |  emd-10430.cif.gz emd-10430.cif.gz | 9.4 KB | ||
| Others |  emd_10430_additional_1.map.gz emd_10430_additional_1.map.gz emd_10430_additional_2.map.gz emd_10430_additional_2.map.gz emd_10430_additional_3.map.gz emd_10430_additional_3.map.gz emd_10430_additional_4.map.gz emd_10430_additional_4.map.gz | 151.5 MB 154.6 MB 153.3 MB 151.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10430 http://ftp.pdbj.org/pub/emdb/structures/EMD-10430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10430 | HTTPS FTP |
-Related structure data
| Related structure data |  6tayMC  6taxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10334 (Title: Cryo-EM structure of RNF213 reveals a RING-type E3 with a dynein core and cysteine reactivity EMPIAR-10334 (Title: Cryo-EM structure of RNF213 reveals a RING-type E3 with a dynein core and cysteine reactivityData size: 1.9 TB Data #1: unaligned multi-frame micrographs of moyamoya variant of RNF213 (R4753K) [micrographs - multiframe] Data #2: unaligned multi-frame micrographs of wildtype RNF213 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10430.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10430.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map assembled from 4 focused refinements | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
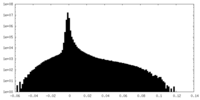
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
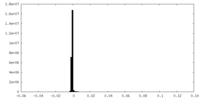
-Additional map: None
| File | emd_10430_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
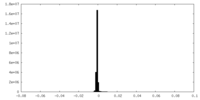
| Projections & Slices |
| ||||||||||||
| Density Histograms |
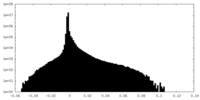
-Additional map: focused refinement on the second half of the AAA-region
| File | emd_10430_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement on the second half of the AAA-region | ||||||||||||
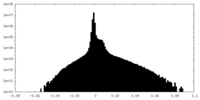
| Projections & Slices |
| ||||||||||||
| Density Histograms |
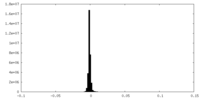
-Additional map: None
| File | emd_10430_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
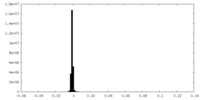
-Additional map: focused refinement on the N-terminus
| File | emd_10430_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement on the N-terminus | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : full length mouse RNF213 with the R4753K mutation
| Entire | Name: full length mouse RNF213 with the R4753K mutation |
|---|---|
| Components |
|
-Supramolecule #1: full length mouse RNF213 with the R4753K mutation
| Supramolecule | Name: full length mouse RNF213 with the R4753K mutation / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 580 KDa |
-Macromolecule #1: RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein li...
| Macromolecule | Name: RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 527.650312 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)KLTLGLSIL FMVEAAEFTV PKKDLDSLCY LLIPSAGSPE ALHSDLSPVL RIRQRWRIYL TNLCLR CID ERCDRWLGIL PLLHTCMQKS PPKKNSKSQP EDTWAGLEGI SFSEFRDKAP TRSQPLQFMQ SKMALLRVDE YLFRSWL SV VPLESLSSYL ENSIDYLSDV PVRVLDCLQG ISYRLPGLRK ISNQNMKKDV ENVFKMLMHL VDIYQHRIFG ENLLQIYL T ECLTLHETVC NITANHQFFE IPALSAELIC KLLELSPPGH TDEGLPEKSY EDLVTSTLQE ALATTRNWLR SLFKSRMLS ISSAYVRLTY SEEMAVWRRL VEIGFPEKHG WKGSLLGDME GRLKQEPPRL QISFFCSSQC RDGGLHDSVS RSFEKCVIEA VSSACQSQT SVLEGLSCQD LQKFGTLLSA VITKSWPVHN GEPVFDVDEI FKYLLKWPDV RQLFELCGTN EKIIDNITEE G RQLMATAE SVFQKVAGEL ENGTIVVGQL ELILEHQSQF LDIWNLNRRR LPSQEKACDV RSLLKRRRDD LLFLKQEKRY VE SLLRQLG RVKHLVQVDF GNIEIIHSQD LSNKKLNEAV IKLPNSSSYK RETHYCLSPD IREMASKLDS LKDSHIFQDF WQE TAESLN TLDKDPRELK VSLPEVLEYL YNPCYDNFYT LYENLKSGKI TFAEVDAIFK DFVDKYDELK NDLKFMCTMN PQDQ KGWIS ERVGQIKEYH TLHQAVSSAK VILQVRRALG VTGDFSVLNP LLNFADSFED FGNEKLDQIS PQFIKAKQLL QDISE PRQR CLEELARQTE LVAWLHKALE DINELKVFVD LASISAGEND IDVDRVACFH DAVQGYASLL YKMDERTNFS DFMNHL QEL WRALDNDQHL PDKLKDSARN LEWLKTVKES HGSVELSSLS LATAINSRGV YVIEAPKDGQ KISPDTVLRL LLPDGHG YP EALRTYSTEE LKELLNKLML MSGKKDHNSN TEVEKFSEVF SNMQRLVHVF IKLHCAGNML FRTWTAKVYC CPDGGIFM N FGLELLSQLT EKGDVIQLLG ALCRQMEDFL DNWKTVVAQK RAEHFYLNFY TAEQLVYLSS ELRKPRPSEA ALMMLSFIK GKCTVQDLVQ ATSACESKAD RYCLREVMKK LPQQLLSEPS LMGKLQVIMM QSLVYMSAFL PHCLDLDALG RCLAHLATMG GTPVERPLP KGLQAGQPNL ILCGHSEVLP AALAIYMQAP RQPLPTFDEV LLCTPATTIE EVELLLRRCL TSGSQGHKVY S LLFADQLS YEVGCQAEEF FQSLCTRAHR EDYQLVILCD AAREHCYIPS TFSQYKVPLV PQAPLPNIQA YLQSHYQVPK RL LSAATVF RDGLCVGIVT SERAGVGKSL YVNTLHTKLK AKLRDETVPL KIIRLTEPHL DENQVLSALL PFLKEKYQKM PVI FHIDIS TSVQTGIPIF LFKLLILQYL MDINGKIWRR SPGHLYLVEI PQGLSVQPKR SSKLNARAPL FKFLDLFPKV TCRP PKEVI DMELTPERSH TDPAMDPVEF CSEAFQRPYQ YLKRFHQQQN LDTFQYEKGS VEGSPEECLQ HFLIYCGLIN PSWSE LRNF AWFLNCQLKD CEASIFCKSA FTGDTLRGFK NFVVTFMILM ARDFATPTLH TSDQSPGRQS VTIGEVVEED LAPFSL RKR WESEPHPYVF FNGDHMTMTF IGFHLETNNN GYVDAINPSN GKVIKKDVMT KELFDGLRLQ RVPFNIDFDN LPRYEKL ER LCLALGIEWP IDPDETYELT TDNMLKILAI EMRFRCGIPV IIMGETGCGK TRLIKFLSDL KRGSVEAETM KLVKVHGG T TPSMIYSKVK EAERTAFSNK AQHKLDTILF FDEANTTEAV SCIKEILCDR TVDGEHLHED SGLHIIAACN PYRKHSQEM ILRLESAGLG YRVSAEETAD RLGSIPLRQL VYRVHALPPS LIPLVWDFGQ LNDSAEKLYI QQIVQRLVDS VSVNPSETCV IADVLSASQ MFMRKRENEC GFVSLRDVER CVKVFRWFHD HSDMLLKELD KFLHESSDST HTFERDPVLW SLVMAIGVCY H ASLEEKAS YRTAIARCFP KPYNSSRAIL DEVTHVQDLF LRGAPIRTNI ARNLALKENV FMMVICIELK IPLFLVGKPG SS KSLAKII VADAMQGQAA FSELFRCLKQ VHLVSFQCSP HSTPQGIIST FKQCARFQQG KDLGQYVSVV VLDEVGLAED SPK MPLKTL HPLLEDGCIE DDPAPYKKVG FVGISNWALD PAKMNRGIFV SRGSPNEKEL IESAEGICSS DRLVQDKIRG YFAP FAKAY ETVCQKQDKE FFGLRDYYSL IKMVFAKAKA SKRGLSPQDI THAVLRNFSG KDNIQALSIF TASLPEARYK EEVST VELI KQNIYPGPQA SSRGLDGAES RYLLVLTRNY VALQILQQTF FEGQQPEIIF GSSFPQDQEY TQICRNINRV KICMET GKM VVLLNLQNLY ESLYDALNQY YVYLGGQKYV DLGLGTHRVK CRVHTAFRLI VIEEKDVVYK QFPVPLINRL EKHYLDM NT VLQPWQKSIV QELQQWAHEF ADVKADQFIA RHKYSPADVF IGYHSDACAS VVLQAVERQG CRDLTEELYR KVSEEARS I LLDCATPDAV VRLSGSSLGS FTAKQLSQEY YYAQQHNSFV DFLQAHLRMT HHECRAVFTE ITTFSRLLTG NDCDVLASE LRGLASKPVV LSLQQYDTEY SFLKDVRSWL TNPGKRKVLV IQADFDDGTR SAQLVASAKY TAINEINKTQ GTKDFVFVYF VTKLSRMGS GTSYVGFHGG LWRSVHIDDL RRSTIMASDV TKLQNVTISQ LFKPEDKPEQ EEMEIETSQS KELAEEQMEV E DSEEMKKA SDPRSCDCSQ FLDTTRLVQS CVQGAVGMLR DQNESCARNM RRVTILLDLL NEDNTRNASF LRESKMRLHV LL NKQEENQ VRSLKEWVTR EAANQDALQE AGTFRHTLWK RVQDVVTPIL ASMIAHIDRD GNLELLAQPD SPAWVQDLWM FIY SDIKFL NISLVLNNTR SNSEMSFILV QSHMNLLKDA YNAVPFSWRI RDYLEELWVQ AQYITDTEGL SKKFVEIFQK TPLG VFLAQ FPVAQQQKLL QSYLKDFLLL TMKVSSREEL MFLQMALWSC LRELQEASGT PDETYKFPLS LPWVHLAFQH FRTRL QNFS RILTIHPQVL SSLSQAAEKH SLAGCEMTLD AFAAMACAEM LKGDLLKPSP KAWLQLVKNL STPLELVCSE GYLCDS GSM TRSVIQEVRA LWNRIFSIAL FVEHVLLGTE SHIPELSPLV TTYVSLLDKC LEEDSNLKTC RPFVAVMTTL CDCKDKA SK KFSRFGIQPC FICHGDAQDP VCLPCDHVYC LRCIQTWLIP GQMMCPYCLT DLPDKFSPTV SQDHRKAIEK HAQFRHMC N SFFVDLVSTM CFKDNTPPEK SVIDTLLSLL FVQKELLRDA SQKHREHTKS LSPFDDVVDQ TPVIRSVLLK LLLKYSFHE VKDYIQNYLT QLEKKAFLTE DKTELYLLFI SCLEDSVHQK TSAGCRNLEQ VLREEGHFLR TYSPGLQGQE PVRIASVEYL QEVARVRLC LDLAADFLSE LQEGSELAED KRRFLKHVEE FCTRVNNDWH RVYLVRKLSS QRGMEFVQSF SKQGHPCQWV F PRKVIAQQ KDHVSLMDRY LVHGNEYKAV RDATAKAVLE CKTLDIGNAL MACRSPKPQQ TAYLLLALYT EVAALYRSPN GS LHPEAKQ LEAVNKFIKE SKILSDPNIR CFARSLVDNT LPLLKIRSAN SILKGTVTEM AVHVATILLC GHNQILKPLR NLA FYPVNM ANAFLPTMPE DLLVHARTWR GLENVTWYTC PRGHPCSVGE CGRPMQESTC LDCGLPVGGL NHTPHEGFSA IRNN EDRTQ TGHVLGSPQS SGVAEVSDRG QSPVVFILTR LLTHLAMLVG ATHNPQALTV IIKPWVQDPQ GFLQQHIQRD LEQLT KMLG RSADETIHVV HLILSSLLRV QSHGVLNFNA ELSTKGCRNN WEKHFETLLL RELKHLDKNL PAINALISQD ERISSN PVT KIIYGDPATF LPHLPQKSII HCSKIWSCRR KITVEYLQHI VEQKNGKETV PVLWHFLQKE AELRLVKFLP EILALQR DL VKQFQNVSRV EYSSIKGFIH SHSSDGLRKL LHDRITIFLS TWNALRRSLE TNGEIKLPKD YCCSDLDLDA EFEVILPR R QGLGLCGTAL VSYLISLHNN MVYTVQKFSN EDNSYSVDIS EVADLHVISY EVERDLNPLI LSNCQYQVQQ GGETSQEFD LEKIQRQISS RFLQGKPRLT LKGIPTLVYR RDWNYEHLFM DIKNKMAQSS LPNLAISTIS GQLQSYSDAC EALSIIEITL GFLSTAGGD PGMDLNVYIE EVLRMCDQTA QVLKAFSRCQ LRHIIALWQF LSAHKSEQRL RLNKELFREI DVQYKEELST Q HQRLLGTF LNEAGLDAFL LELHEMIVLK LKGPRAANSF NPNWSLKDTL VSYMETKDSD ILSEVESQFP EEILMSSCIS VW KIAATRK WDRQSRGGGH HHHHHHHHH UniProtKB: E3 ubiquitin-protein ligase RNF213 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.01 kPa Details: SCD 005 Sputter Coater (BAL-TEC) grid sitting on a metallic mesh during glow discharge | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP Details: blot for 1.5 seconds using using Whatman Filter Paper Grade 1. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 6561 / Average exposure time: 10.0 sec. / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6tay: |
 Movie
Movie Controller
Controller









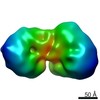




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)