[English] 日本語
 Yorodumi
Yorodumi- PDB-1d8e: Zinc-depleted FTase complexed with K-RAS4B peptide substrate and ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1d8e | ||||||
|---|---|---|---|---|---|---|---|
| Title | Zinc-depleted FTase complexed with K-RAS4B peptide substrate and FPP analog. | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE / FTASE / PFT / PFTASE / FARNESYLTRANSFERASE / FARNESYL TRANSFERASE / CAAX / RAS / CANCER | ||||||
| Function / homology |  Function and homology information Function and homology informationApoptotic cleavage of cellular proteins / Inactivation, recovery and regulation of the phototransduction cascade / RAS processing / protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase complex / protein farnesylation / protein geranylgeranyltransferase type I / positive regulation of tubulin deacetylation / protein farnesyltransferase ...Apoptotic cleavage of cellular proteins / Inactivation, recovery and regulation of the phototransduction cascade / RAS processing / protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase complex / protein farnesylation / protein geranylgeranyltransferase type I / positive regulation of tubulin deacetylation / protein farnesyltransferase / protein farnesyltransferase activity / protein farnesyltransferase complex / Rab geranylgeranyltransferase activity / regulation of fibroblast proliferation / protein geranylgeranylation / positive regulation of skeletal muscle acetylcholine-gated channel clustering / : / farnesyltranstransferase activity / microtubule associated complex / forebrain astrocyte development / negative regulation of epithelial cell differentiation / regulation of synaptic transmission, GABAergic / type I pneumocyte differentiation / epithelial tube branching involved in lung morphogenesis / enzyme-linked receptor protein signaling pathway / Rac protein signal transduction / : / skeletal muscle cell differentiation / positive regulation of Rac protein signal transduction / Signaling by RAS GAP mutants / Signaling by RAS GTPase mutants / Activation of RAS in B cells / RAS signaling downstream of NF1 loss-of-function variants / RUNX3 regulates p14-ARF / SOS-mediated signalling / Activated NTRK3 signals through RAS / Activated NTRK2 signals through RAS / SHC1 events in ERBB4 signaling / alpha-tubulin binding / Signalling to RAS / SHC-related events triggered by IGF1R / Activated NTRK2 signals through FRS2 and FRS3 / glial cell proliferation / Estrogen-stimulated signaling through PRKCZ / SHC-mediated cascade:FGFR3 / MET activates RAS signaling / Signaling by PDGFRA transmembrane, juxtamembrane and kinase domain mutants / Signaling by PDGFRA extracellular domain mutants / SHC-mediated cascade:FGFR2 / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / positive regulation of cell cycle / SHC-mediated cascade:FGFR4 / Signaling by FGFR4 in disease / Erythropoietin activates RAS / SHC-mediated cascade:FGFR1 / protein-membrane adaptor activity / FRS-mediated FGFR3 signaling / Signaling by CSF3 (G-CSF) / Signaling by FLT3 ITD and TKD mutants / positive regulation of glial cell proliferation / FRS-mediated FGFR2 signaling / FRS-mediated FGFR4 signaling / Signaling by FGFR3 in disease / p38MAPK events / homeostasis of number of cells within a tissue / Tie2 Signaling / FRS-mediated FGFR1 signaling / Signaling by FGFR2 in disease / striated muscle cell differentiation / GRB2 events in EGFR signaling / SHC1 events in EGFR signaling / EGFR Transactivation by Gastrin / Signaling by FLT3 fusion proteins / FLT3 Signaling / Signaling by FGFR1 in disease / GRB2 events in ERBB2 signaling / CD209 (DC-SIGN) signaling / Ras activation upon Ca2+ influx through NMDA receptor / NCAM signaling for neurite out-growth / SHC1 events in ERBB2 signaling / Downstream signal transduction / Constitutive Signaling by Overexpressed ERBB2 / Insulin receptor signalling cascade / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / small monomeric GTPase / VEGFR2 mediated cell proliferation / response to cytokine / FCERI mediated MAPK activation / Signaling by ERBB2 TMD/JMD mutants / RAF activation / wound healing / Constitutive Signaling by EGFRvIII / regulation of long-term neuronal synaptic plasticity / Signaling by high-kinase activity BRAF mutants / Signaling by ERBB2 ECD mutants / MAP2K and MAPK activation / visual learning / Signaling by ERBB2 KD Mutants / Signaling by SCF-KIT / lipid metabolic process Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 3 Å X-RAY DIFFRACTION / Resolution: 3 Å | ||||||
 Authors Authors | Long, S.B. / Casey, P.J. / Beese, L.S. | ||||||
 Citation Citation |  Journal: Structure Fold.Des. / Year: 2000 Journal: Structure Fold.Des. / Year: 2000Title: The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures. Authors: Long, S.B. / Casey, P.J. / Beese, L.S. #1:  Journal: Science / Year: 1997 Journal: Science / Year: 1997Title: Crystal Structure of Protein Farnesyltransferase at 2.25A Resolution Authors: Park, H.-W. / Boduluri, S.R. / Moomaw, J.F. / Casey, P.J. / Beese, L.S. #2:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Co-crystal Structure of Mammalian Protein Farnesyltransferase with a Farnesyl Diphosphate Substrate Authors: Long, S.B. / Casey, P.J. / Beese, L.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1d8e.cif.gz 1d8e.cif.gz | 161.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1d8e.ent.gz pdb1d8e.ent.gz | 126.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1d8e.json.gz 1d8e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1d8e_validation.pdf.gz 1d8e_validation.pdf.gz | 664.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1d8e_full_validation.pdf.gz 1d8e_full_validation.pdf.gz | 675.4 KB | Display | |
| Data in XML |  1d8e_validation.xml.gz 1d8e_validation.xml.gz | 28.9 KB | Display | |
| Data in CIF |  1d8e_validation.cif.gz 1d8e_validation.cif.gz | 40.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d8/1d8e https://data.pdbj.org/pub/pdb/validation_reports/d8/1d8e ftp://data.pdbj.org/pub/pdb/validation_reports/d8/1d8e ftp://data.pdbj.org/pub/pdb/validation_reports/d8/1d8e | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-FARNESYLTRANSFERASE ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 44098.145 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein | Mass: 48722.281 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Protein/peptide , 1 types, 1 molecules P
| #3: Protein/peptide | Mass: 1298.723 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: This synthetic peptide is derived from human K-Ras4B, residues 178-188 References: UniProt: P01116 |
|---|
-Non-polymers , 3 types, 77 molecules 
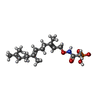



| #4: Chemical | ChemComp-ACT / |
|---|---|
| #5: Chemical | ChemComp-FII / [( |
| #6: Water | ChemComp-HOH / |
-Details
| Compound details | The peptide adopts a non-catalytic beta-turn conformation in the absence of zinc. Please refer to ...The peptide adopts a non-catalytic beta-turn conformation in the absence of zinc. Please refer to the 2.0A resolution structure in the presence of zinc, 1D8D. residues 1-54 of chain A are disordered residues 1-16 and 424-437 of chain b are disordered residues 1-5 of chain P are disordered |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.14 Å3/Da / Density % sol: 60.79 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 290 K / Method: vapor diffusion, hanging drop / pH: 5.7 Details: Peg 8000, ammonium acetate, DTT , pH 5.7, VAPOR DIFFUSION, HANGING DROP, temperature 290K | ||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 17 ℃ | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 96 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: Mar 5, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 3→50 Å / Num. all: 22662 / Num. obs: 21199 / % possible obs: 89.3 % / Observed criterion σ(I): 0 / Redundancy: 4.01 % / Rmerge(I) obs: 0.09 / Net I/σ(I): 7.9 |
| Reflection shell | Resolution: 3→3.03 Å / Redundancy: 2.4 % / Rmerge(I) obs: 0.289 / Num. unique all: 770 / % possible all: 96 |
| Reflection | *PLUS Num. measured all: 85082 |
| Reflection shell | *PLUS % possible obs: 82.2 % / Mean I/σ(I) obs: 2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 3→50 Å / Rfactor Rfree error: 0.008 / Data cutoff high absF: 10000000 / Data cutoff low absF: 0.001 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 2 / σ(I): -3 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.8 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→50 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3→3.14 Å / Rfactor Rfree error: 0.037 / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller















 PDBj
PDBj






















