+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9944 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human MCU-EMRE complex | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ion channel / Mitochondrial Ca2+ uptake / MCU / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationuniporter activity / mitochondrial calcium ion transmembrane transport / uniplex complex / Processing of SMDT1 / positive regulation of mitochondrial calcium ion concentration / Mitochondrial calcium ion transport / mitochondrial calcium ion homeostasis / calcium import into the mitochondrion / channel activator activity / cellular response to calcium ion starvation ...uniporter activity / mitochondrial calcium ion transmembrane transport / uniplex complex / Processing of SMDT1 / positive regulation of mitochondrial calcium ion concentration / Mitochondrial calcium ion transport / mitochondrial calcium ion homeostasis / calcium import into the mitochondrion / channel activator activity / cellular response to calcium ion starvation / positive regulation of neutrophil chemotaxis / positive regulation of mitochondrial fission / protein complex oligomerization / calcium channel complex / Mitochondrial protein degradation / calcium-mediated signaling / positive regulation of insulin secretion / calcium channel activity / glucose homeostasis / protein-macromolecule adaptor activity / mitochondrial inner membrane / mitochondrial matrix / mitochondrion / nucleoplasm / identical protein binding Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | ||||||||||||
 Authors Authors | Zhuo W / Zhou H | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2021 Journal: Protein Cell / Year: 2021Title: Structure of intact human MCU supercomplex with the auxiliary MICU subunits. Authors: Wei Zhuo / Heng Zhou / Runyu Guo / Jingbo Yi / Laixing Zhang / Lei Yu / Yinqiang Sui / Wenwen Zeng / Peiyi Wang / Maojun Yang /  | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9944.map.gz emd_9944.map.gz | 8.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9944-v30.xml emd-9944-v30.xml emd-9944.xml emd-9944.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9944.png emd_9944.png | 85.6 KB | ||
| Filedesc metadata |  emd-9944.cif.gz emd-9944.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9944 http://ftp.pdbj.org/pub/emdb/structures/EMD-9944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9944 | HTTPS FTP |
-Related structure data
| Related structure data |  6k7xMC  9945C  6k7yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9944.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9944.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
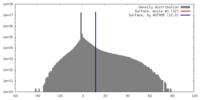
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human MCU-EMRE complex
| Entire | Name: Human MCU-EMRE complex |
|---|---|
| Components |
|
-Supramolecule #1: Human MCU-EMRE complex
| Supramolecule | Name: Human MCU-EMRE complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Calcium uniporter protein, mitochondrial
| Macromolecule | Name: Calcium uniporter protein, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.300203 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DDVTVVYQNG LPVISVRLPS RRERCQFTLK PISDSVGVFL RQLQEEDRGI DRVAIYSPDG VRVAASTGID LLLLDDFKLV INDLTYHVR PPKRDLLSHE NAATLNDVKT LVQQLYTTLC IEQHQLNKER ELIERLEDLK EQLAPLEKVR IEISRKAEKR T TLVLWGGL ...String: DDVTVVYQNG LPVISVRLPS RRERCQFTLK PISDSVGVFL RQLQEEDRGI DRVAIYSPDG VRVAASTGID LLLLDDFKLV INDLTYHVR PPKRDLLSHE NAATLNDVKT LVQQLYTTLC IEQHQLNKER ELIERLEDLK EQLAPLEKVR IEISRKAEKR T TLVLWGGL AYMATQFGIL ARLTWWEYSW DIMEPVTYFI TYGSAMAMYA YFVMTRQEYV YPEARDRQYL LFFHKGAKKS RF DLEKYNQ LKDAIAQAEM DLKRLRDPLQ VHLPLRQIG UniProtKB: Calcium uniporter protein, mitochondrial |
-Macromolecule #2: Essential MCU regulator, mitochondrial
| Macromolecule | Name: Essential MCU regulator, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.993192 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VIVTRSGAIL PKPVKMSFGL LRVFSIVIPF LYVGTLISKN FAALLEEHDI FVPE UniProtKB: Essential MCU regulator, mitochondrial |
-Macromolecule #3: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 3 / Number of copies: 8 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Macromolecule #4: (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,...
| Macromolecule | Name: (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL type: ligand / ID: 4 / Number of copies: 16 / Formula: PLX |
|---|---|
| Molecular weight | Theoretical: 767.132 Da |
| Chemical component information |  ChemComp-PLX: |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.27 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 179468 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6k7x: |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)